Abstract
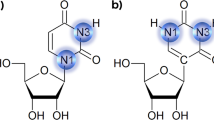
The dynamic structure of 11-mer DNA duplexes of different sequences with or without homopyrimidine (T·T, or BrdU·T) mismatches was studied by molecular dynamics (MD) simulations on a time scale from 200 ps to 1 ns. The conformational analysis suggests that in mismatched duplexes the formation of classical T·T wobble H-bonding pairing is nearest-neighbor sequence-dependent and, in most cases, three-centered H-bonds and numerous alternative close cross-strand interatomic contacts exist. Thus, in duplex W1, where the central triplet is 5′d(CTA)·d(TTG), two wobble conformations W↑ (αβ) and W↓ (βα) are formed and exchange rapidly at 300 K. In contrast, when the central triplet is 5′d(TTT)·d(ATA) (W2 duplex) wobble conformations are rarely observed at 300 K, and the T·T mispair most often adopts a “twisted” conformation with one largely persistent normal H-bond, plus a stable cross-strand contact involving a T flanking base. However, at elevated temperature (400 K) the same W2 duplex shows frequent exchange between the two classical wobble conformations (αβ↔βα), as is in the case when the central triplet is 5′d(TBrdUT)·d(ATA) (W3 duplex at 300 K). It is suggested that in the W2 sequence, restrictions due to thymine-methyl/π interactions prevent the formation of wobble pairing and thermal activation energy, and/or the chemical replacement of T by BrdU are required in order for the T(BrdU)·T mismatch to adopt and exchange between wobble conformations. The specific short and/or long-lived (double/triple) cross-strand dynamic interactions in W1, W2 and W3 duplexes are throughout characterized. These frequent atomic encounters exemplify possible inter-strand charge transfer pathways in the studied DNA molecules.
Figure 3D structure snapshots of wobble and frequent overlapping conformers formed within the W3 central triplet during 200 ps MD: α’+ ϒ’. H-bonds (magenta) and close cross-strand contacts, Å (orange).















Similar content being viewed by others
References
Brown T, Kennard O, Kneale G, Rabinovich D (1985) Nature 315:604–606
Brown T, Kneale G, Hunter WN, Kennard O (1986) Nucl Acids Res 14:1801–1809
Privé GG, Heinemann U, Chandrasegaran S, Kan L-S, Kopka ML, Dickerson RE (1987) Science 238:498–504
Maskos K, Gunn MB, LeBlanc DA, Morden KM (1993) Biochemistry 32:3583–3595
Boulard Y, Cognet JAH, Gabarro-Arpa J, Le Bret M, Carbonnaux C, Fazakerley GV (1995) J Mol Biol 246:194–208
Boulard Y, Fazakerley GV, Sowers LC (2002) Nucl Acids Res 30:1371–1378
Isaacs RJ, Rayens WS, Spielmann HP (2002) J Mol Biol 319:191–207
Venable RM, Widmalm G, Brooks BR, Egan W, Pastor RW (1992) Biopolymers 32:783–794
Špačková N, Berger I, Šponer J (2000) J Am Chem Soc 122:7564–7572
Limoli CL, Ward JF (1993) Radiat Res 134:160–169
Gralla J, Crothers DM (1973) J Mol Biol 78:301–319
Peyret N, Seneviratne PA, Allawi HT, SantaLucia Jr J (1999) Biochemistry 38:3468–3477
Robinson BH, Mailer C, Drobny G (1997) Annu Rev Biophys Biomol Struct 26:629–658
Swaminathan S, Ravishanker G, Beveridge DL (1991) J Am Chem Soc 113:5027–5040
McAteer K, Kennedy MA (2003) J Biomol Struct Dynam 20:487–506
Sowers LC, Goodman MF, Eritja R, Kaplan B, Fazakerley GV (1989) J Mol Biol 205:437–447
Arnold FH, Wolk S, Cruz P, Tinoco Jr I (1987) Biochemistry 26:4068–4075
Kouchakdjian M, Li BFL, Swann PF, Patel DJ (1988) J Mol Biol 202:139–155
Gervais V, Cognet JAH, Le Bret M, Sowers LC, Fazakerley GV (1995) Eur J Biochem 228:279–290
Yoon C, Privé GG, Goodsell DS, Dickerson RE (1988) Proc Natl Acad Sci USA 85:6332–6336
Sommers MF, Byrd RA, Gallo KA, Samson CJ, Zon G, Egan W (1985) Nucl Acids Res 13:6375–6386
Cecchini S, Girouard S, Huels MA, Sanche L, Hunting DJ (2004) Radiat Res 162:604--615
Myers RM, Fischer SG, Maniatis T, Lerman LS (1985) Nucl Acids Res 13:3111–3129
Arnott S, Hukins DWL (1972) Biochem Biophys Res Comm 47:1504–1509
Boulard Y, Cognet JAH, Fazakerley GV (1997) J Mol Biol 268:331–347
Cognet JAH, Gabarro-Arpa, Cuniasse Ph, Fazakeley GV, Le Bret M (1990) J Biomol Struct Dynam 7:1095–1115
Boulard Y, Cognet JAH, Gabarro-Arpa J, Le Bret M, Sowers LC, Fazakerley GV (1992) Nucl Acids Res 20:1933–1941
Cognet JAH, Boulard Y, Fazakerley GV (1995) J Mol Biol 246:209–226
Lavery R, Sklenar H (1989) J Biomol Struct Dynam 6:655–667
Real AN, Greenall RJ (2000) J Mol Model 6:654–658
Umezawa Y, Nishio M (2002) Nucl Acids Res 30:2183–2192
Marcus RA, Sutin N (1985) Biochim Biophys Acta 811:265–322
Acknowledgements
This work was supported by the NCI of Canada. Supplementary material: BrdU AM1 structure, MM optimized and MD DNA 3D-structures (mol2 and pdb formats), dynamics history files, etc. are available on request.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gantchev, T.G., Cecchini, S. & Hunting, D.J. Dynamic conformational states of DNA containing T·T or BrdU·T mispaired bases: wobble H-bond pairing versus cross-strand inter-atomic contacts. J Mol Model 11, 141–159 (2005). https://doi.org/10.1007/s00894-005-0238-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-005-0238-9