Abstract
Artificial neural networks (ANNs) have been successfully trained to model and predict the acidity constants (pK a) of 128 various phenols with diverse chemical structures using a quantitative structure-activity relationship. An ANN with 6-14-1 architecture was generated using six molecular descriptors that appear in the multi-parameter linear regression (MLR) model. The polarizability term (π I), most positive charge of acidic hydrogen atom (q +), molecular weight (MW), most negative charge of the phenolic oxygen atom (q −), the hydrogen-bond accepting ability (ɛ B) and partial-charge weighted topological electronic (PCWTE) descriptors are inputs and its output is pK a. It was found that a properly selected and trained neural network with 106 phenols could represent the dependence of the acidity constant on molecular descriptors fairly well. For evaluation of the predictive power of the ANN, an optimized network was used to predict the pK as of 22 compounds in the prediction set, which were not used in the optimization procedure. A squared correlation coefficient (R 2) and root mean square error (RMSE) of 0.8950 and 0.5621 for the prediction set by the MLR model should be compared with the values of 0.99996 and 0.0114 by the ANN model. These improvements are due to the fact that the pK a of phenols shows non-linear correlations with the molecular descriptors.

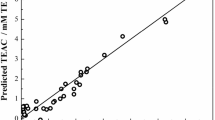
Plot of the calculated values of pK a from the ANN model versus the experimental values of it for training, validation and prediction sets.





Similar content being viewed by others
References
Cronce DT, Famini GR, Soto JAD, Wilson LY (1998) J Chem Soc Perkin Trans 2:1293–1301
Engberts JBFN, Famini GR, Perjessy A, Wilson LY (1998) J Phys Org Chem 11:261–272
McClelland HE, Jurs PC (2000) J Chem Inf Comput Sci 40:967–975
Hiob R, Karelson M (2000) J Chem Inf Comut Sci 40:1062–1071
Habibi-Yangjeh A (2004) Indian J Chem B 43:1504–1526
Ma Y, Gross KC, Hollingsworth CA, Seybold PG, Murray JS (2004) J Mol Model 10:235–239
Karelson M, Lobanov VS (1996) Chem Rev 96:1027–1043
Todeschini R, Consonni V (2000) Handbook of Molecular Descriptors. Wiley-VCH, Weinheim, Germany
Kramer R (1998) Chemometric techniques for quantitative analysis. Marcel Dekker, New York
Kuzmanovski I, Aleksovska S (2003) Chemom Intell Lab Syst 67:167–174
Barros AS, Rutledge DN (1998) Chemomet Intell Lab Syst 40:65–72
Garkani-Nejad Z, Karlovits M, Demuth W, Stimpfl T, Vycudilik W, Jalali-Heravi M, Varmuza K (2004) J Chromatogr A 1028:287–295
Patterson DW (1996) Artificial neural networks: theory and applications. Simon and Schuster, New York
Zupan J, Gasteiger J (1999) Neural networks in chemistry and drug design. Wiley-VCH, Weinheim
Agatonovic-Kustrin S, Beresford R (2000) J Pharm Biomed Anal 22:717–727
Fatemi MH (2002) J Chromatogr A 955:273–280
Bunz AP, Braun B, Janowsky R (1999) Fluid Phase Equilib 158:367–374
Homer J, Generalis SC, Robson JH (1999) Phys Chem Chem Phys 1:4075–4081
Urata S, Takada A, Uchimaru T, Chandra AK, Sekiya A (2002) J Fluorine Chem 116:163–171
Koziol J (2002) Internet Electron J Mol Des 1:80–93
Habibi-Yangjeh A, Nooshyar M (2005) Bull Korean Chem Soc 26:139–145
Habibi-Yangjeh A, Nooshyar M (2005) Physics and Chemistry of Liquids 43:239–247
Jalali-Heravi M, Masoum S, Shahbazikhah P (2004) J Magn Reson 171:176–185
Selassie CD, DeSoyza TV, Rosario M, Gao H, Hansch C (1998) Chemico-Biological Interaction 113:175–182
Gruber C, Buss V (1989) Chemosphere 19:1595–1609
Citra MJ (1999) Chemosphere 38:191–206
Schuurmann G (1996) Quant Struct Act Relat 15:121–132
Gross KC, Seybold PG (2001) Int J Quant Chem 85:569–579
Liptak MD, Gross KC, Seybold PG, Feldgus S, Shields GC (2002) J Am Chem Soc 124:6421–6427
Hanai T, Koizumi K, Kinoshita T (2000) J Liq Chromatogr Relat Technol 23:363–385
HyperChem, Release 7.0 for Windows (2002) Molecular Modeling System, Hypercube Inc
Todeschini R, Consonni V, Pavan M (2002) Dragon Software Version 2.1
Dean JA (1999) Lange’s Handbook of Chemistry, 15th edn. McGraw-Hill Inc.
Demuth H, Beale M (2000) Neural network toolbox. Mathworks, Natick MA
Despagne F, Massart DL (1998) Analyst 123:157R–178R
Matlab 6.5. (1984–2002) Mathworks
Famini GR, Wilson LY (1999) J Phys Org Chem 12:645–652
Acknowledgement
The Authors wish to acknowledge the vice-presidency of research, university of Mohaghegh Ardebili, for financial support of this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Habibi-Yangjeh, A., Danandeh-Jenagharad, M. & Nooshyar, M. Application of artificial neural networks for predicting the aqueous acidity of various phenols using QSAR. J Mol Model 12, 338–347 (2006). https://doi.org/10.1007/s00894-005-0050-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-005-0050-6