Abstract
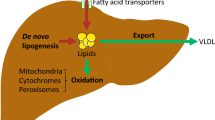
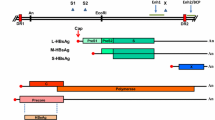
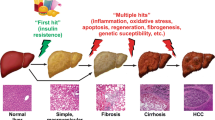
Hepatitis C virus (HCV) infection has been known to use autophagy for its replication. However, the mechanisms by which HCV modulates autophagy remain controversial. We used HCV-Japanese fulminant hepatitis-1-infected Huh7 cells. HCV infection induced the accumulation of autophagosomes. Morphological analyses of monomeric red fluorescent protein (mRFP)-green fluorescent protein (GFP) tandem fluorescent-tagged LC3 transfection showed HCV infection impaired autophagic flux. Autophagosome-lysosome fusion assessed by transfection of mRFP- or GFP-LC3 and immunostaining of lysosomal-associated membrane protein 1 was inhibited by HCV infection. Decrease of HCV-induced endoplasmic reticulum (ER) stress by 4-phenylbutyric acid, a chemical chaperone, improved the HCV-mediated autophagic flux impairment. HCV infection-induced oxidative stress and subsequently DNA damage, but not apoptosis. Furthermore, HCV induced cytoprotective effects against the cellular stress by facilitating the formation of cytoplasmic inclusion bodies as shown by p62 expression and by modulating keratin protein expression and activated nuclear factor erythroid 2-related factor 2. HCV eradication by direct-acting antivirals improved autophagic flux, but DNA damage persisted. In conclusion, HCV-induced ER stress correlates with autophagic flux impairment. Decrease of ER stress is considered to be a promising therapeutic strategy for HCV-related chronic liver diseases. However, we should be aware that the risk of hepatocarcinogenesis remains even after HCV eradication.







Similar content being viewed by others
Abbreviations
- HCV:
-
Hepatitis C virus
- HCC:
-
Hepatocellular carcinoma
- ER:
-
Endoplasmic reticulum
- JFH1:
-
Japanese fulminant hepatitis-1
- NS:
-
Nonstructural
- IFN:
-
Interferon
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- 4PBA:
-
4-Phenylbutyric acid
- 3MA:
-
3-Methyladenine
- SOF:
-
Sofosbuvir
- DCV:
-
Daclatasvir
- DMSO:
-
Dimethyl sulfoxide
- K:
-
Keratin
- CHOP:
-
C/EBP homologous protein
- XBP1:
-
X-box binding protein 1
- SREBP1c:
-
Sterol regulatory element binding protein 1c
- Nrf2:
-
Nuclear factor erythroid 2-related factor 2
- HNE:
-
Hydroxyl-2-nonenal
- γ-H2AX:
-
Gamma-H2AX
- 53BP1:
-
p53-binding protein 1
- Atg:
-
Autophagy-related gene
- SOD1:
-
Superoxide dismutase 1
- eIF2α:
-
Eukaryotic initiation factor 2α
- JNK:
-
c-Jun N-terminal kinase
- CREB:
-
cAMP response element-binding protein
- PARP:
-
Poly-ADP-ribose-polymerase
- LC3:
-
Microtubule-associated protein light chain 3
- Keap1:
-
Kelch-like ECH-associated protein 1
- HO-1:
-
Heme oxygenase-1
- Rubicon:
-
RUN domain and cysteine-rich domain containing, Beclin1-interacting protein
- Lamp1:
-
Lysosomal-associated membrane protein 1
- PFA:
-
Paraformaldehyde
- PBS:
-
Phosphate-buffered saline
- PI:
-
Propidium iodide
- mRFP:
-
Monomeric red fluorescent protein
- GFP:
-
Green fluorescent protein
- tf-LC3:
-
Tandem fluorescent-tagged LC3
- ROS:
-
Reactive oxygen species
- H2DCFDA:
-
2′, 7′-Dichlorodihydrofluorescein diacetate
- MDB:
-
Mallory-Denk body
- DAAs:
-
Direct-acting antivirals
- PI3K:
-
Phosphoinositide 3-kinase
References
Moradpour D, Penin F, Rice CM (2007) Replication of hepatitis C virus. Nat Rev Microbiol 5:453–463
Lok AS, Seeff LB, Morgan TR, di isceqlie AM, Sterling RK, Curto TM, Everson GT, Lindsay KL, Lee WM, Bonkovsky HL, Dienstag JL, Ghany MG, Morishima C, Goodman ZD; HALT-C Trial Group (2009) Incidence of hepatocellular carcinoma and associated risk factors in hepatitis C-related advanced liver disease. Gastroenterology 136:138-148
Mizushima N, Yoshimori T (2007) How to interpret LC3 immunoblotting. Autophagy 3:542–545
Kaizuka T, Morishita H, Hama Y, Tsukamoto S, Matsui T, Toyota Y, Kodama A, Ishihara T, Mizushima T, Mizushima N (2016) An autophagic flux probe that releases an internal control. Mol Cell 64:835–849
Sir D, Kuo CF, Tian Y, Liu HM, Huang EJ, Jung JU, Machida K, Ou JH (2012) Replication of hepatitis C virus RNA on autophagosomal membranes. J Biol Chem 287:18036–18043
Fahmy AM, Labonté P (2017) The autophagy elongation complex (ATG5-12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation. Sci Rep 7:40351
Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M, Bartenschlager R, Liang TJ (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796
Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV (2005) Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA 102:9294–9299
Hara Y, Yanatori I, Ikeda M, Kiyokage E, Nishina S, Toida K, Kishi F, Kato N, Imamura M, Chayama K, Hino K (2014) Hepatitis C virus core protein suppresses mitophagy by interacting with parkin in the context of mitochondrial depolarization. Am J Pathol 184:3026–3039
Kato N, Mori K, Abe K, Dansako H, Kuroki M, Ariumi Y, Wakita T, Ikeda M (2009) Efficient replication systems for hepatitis C virus using a new human hepatoma cell line. Virus Res 146:41–50
Dansako H, Hiramoto H, Ikeda M, Wakita T, Kato N (2014) Rab18 is required for viral assembly of hepatitis C virus through trafficking of the core protein to lipid droplets. Virology 462–463:166–174
Miyagawa K, Oe S, Honma Y, Izumi H, Baba R, Harada M (2016) Lipid-induced endoplasmic reticulum stress impairs selective autophagy at the step of autophagosome-lysosome fusion in hepatocytes. Am J Pathol 186:1861–1873
Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140:313–326
Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y (1991) Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem 266:17707–17712
Harada M, Sakisaka S, Yoshitake M, Kin M, Ohishi M, Shakado S, Mimura Y, Noguchi K, Sata M, Tanikawa K (1996) Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPases, inhibits the receptor-mediated endocytosis of asialoglycoproteins in isolated rat hepatocytes. J Hepatol 24:594–603
Eskelinen EL, Tanaka Y, Saftig P (2003) At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol 13:137–145
Hanada S, Harada M, Kumemura H, Bishr Omary M, Koga H, Kawaguchi T, Taniguchi E, Yoshida T, Hisamoto T, Yanagimoto C, Maeyama M, Ueno T, Sata M (2007) Oxidative stress induces the endoplasmic reticulum stress and facilitates inclusion formation in cultured cells. J Hepatol 47:93–102
Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH (2008) Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054–1061
Kim JY, Wang L, Lee J, Ou JJ (2017) Hepatitis C virus induces the localization of lipid rafts to autophagosomes for its RNA replication. J Virol 91:e00541-e617
Chusri P, Kumthip K, Hong J, Zhu C, Duan X, Jilg N, Fusco DN, Brisac C, Schaefer EA, Cai D, Peng LF, Maneekarn N, Lin W, Chung RT (2016) HCV induces transforming growth factor β1 through activation of endoplasmic reticulum stress and the unfolded protein response. Sci Rep 6:22487
Taguwa S, Kambara H, Fujita N, Noda T, Yoshimori T, Koike K, Moriishi K, Matsuura Y (2011) Dysfunction of autophagy participates in vacuole formation and cell death in cells replicating hepatitis C virus. J Virol 85:13185–13194
Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y (1998) Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 23:33–42
Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T (2009) Two Beclin 1-binding proteins, Atg 14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11:385–396
Wang L, Tian Y, Ou JH (2015) HCV induces the expression of Rubicon and UVRAG to temporally regulate the maturation of autophagosomes and viral replication. PLoS Pathog 11:e1004764
Itakura E, Kishi C, Inoue K, Mizushima N (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19:5360–5372
Shrivastava S, Bhanja Chowdhury J, Steele R, Ray R, Ray RB (2012) Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J Virol 86:8705–9712
Guévin C, Manna D, Bélanger C, Konan KV, Mak P, Labonté P (2010) Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology 405:1–7
Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MM (2011) Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol 85:10561–10571
Kimura S, Noda T, Yoshimori T (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3:452–460
Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465:942–946
Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, Coppes RP, Engedal N, Mari M, Reggiori F (2018) Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14:1435–1455
Seglen PO, Gordon PB (1982) 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA 79:1889–1892
Tsutsumi T, Matsuda M, Aizaki H, Moriya K, Miyoshi H, Fujie H, Shintani Y, Yotsuyanagi H, Miyamura T, Suzuki T, Koike K (2009) Proteomics analysis of mitochondrial proteins reveals overexpression of a mitochondrial protein chaperon, prohibitin, in cells expressing hepatitis C virus core protein. Hepatology 50:378–386
Negro F, Sanyal AJ (2009) Hepatitis C virus, steatosis and lipid abnormalities: clinical and pathogenic data. Liver Int 29(Suppl 2):26–37
Lerat H, Kammoun HL, Hainault I, Mérour E, Higgs MR, Callens C, Lemon SM, Foufelle F, Pawlotsky JM (2009) Hepatitis C virus proteins induce lipogenesis and defective triglyceride secretion in transgenic mice. J Biol Chem 284:33466–33474
Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ (2009) Autophagy regulates lipid metabolism. Nature 458:1131–1135
Vescovo T, Romagnoli A, Perdomo AB, Corazzari M, Ciccosanti F, Alonzi T, Nardacci R, Ippolito G, Tripodi M, Garcia-Monzon C, Lo Iacono O, Piacentini M, Fimia GM (2012) Autophagy protects cells from HCV-induced defects in lipid metabolism. Gastroenterology 142:644–653
Rakoski MO, Brown MB, Fontana RJ, Bonkovsky HL, Brunt EM, Goodman ZD, Lok AS, Omary MB; HALT-C Trial Group (2011) Mallory-Denk bodies are associated with outcomes and histologic features in patients with chronic hepatitis C. Clin Gastroenterol Hepatol 9:902–909
Honma Y, Sato-Morita M, Katsuki Y, Mihara H, Baba R, Harada M (2018) Trehalose activates autophagy and decreases proteasome inhibitor-induced endoplasmic reticulum stress and oxidative stress-mediated cytotoxicity in hepatocytes. Hepatol Res 48:94–105
Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, Cadrin M, Omary MB (2007) From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res 313:2033–2049
Harada M, Strnad P, Resurreccion EZ, Ku NO, Omary MB (2007) Keratin 18 overexpression but not phosphorylation or filament organization blocks mouse Mallory body formation. Hepatology 45:88–96
Kwan R, Hanada S, Harada M, Sternad P, Li DH, Omary MB (2012) Keratin 8 phosphorylation regulates its transamidation and hepatocyte Mallory-Denk body formation. FASEB J 26:2318–2326
Ku NO, Sternad P, Zhong BH, Tao GZ, Omary MB (2007) Keratins let liver live: Mutations predispose to liver disease and crosslinking generates Mallory-Denk bodies. Hepatology 46:1639–1649
Kamegaya Y, Hiasa Y, Zukerberg L, Fowler N, Blackard JT, Lin W, Choe WH, Schmidt EV, Chung RT (2005) Hepatitis C virus acts as a tumor accelerator by blocking apoptosis in a mouse model of hepatocarcinogenesis. Hepatology 41:660–667
Harada M, Hanada S, Toivola DM, Ghori N, Omary MB (2008) Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology 47:2026–2035
Kageyama S, Sou YS, Uemura T, Kametaka S, Saito T, Ishimura R, Kouno T, Bedford L, Mayer RJ, Lee MS, Yamamoto M, Waguri S, Tanaka K, Komatsu M (2014) Proteasome dysfunction activates autophagy and the Keap1-Nrf2 pathway. J Biol Chem 289:24944–24955
Jeong SW, Jang JY, Chung RT (2012) Hepatitis C virus and hepatocarcinogenesis. Clin Mol Hepatol 18:347–356
Lemon SM, McGivern DR (2012) Is hepatitis C virus carcinogenic? Gastroenterology 142:1274–1278
Hino K, Nishina S, Hara Y (2013) Iron metabolic disorder in chronic hepatitis C: mechanisms and relevance to hepatocarcinogenesis. J Gastroenterol Hepatol 28(Suppl 4):93–98
Sharma A, Singh K, Almasan A (2012) Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol Biol 920:613–626
Panier S, Boulton SJ (2014) Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 15:7–18
Siddiqui MS, François M, Fenech MF, Leifert WR (2015) Persistent γH2AX: A promising molecular marker of DNA damage and aging. Mutat Res Rev Mutat Res 766:1–19
Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M (2013) Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 51:618–631
Ioannou GN, Green PK, Berry K (2017) HCV eradication induced by direct-acting antiviral agents reduces the risk of hepatocellular carcinoma. J Hepatol S0168–8287:32273
Waziry R, Hajarizadeh B, Grebely J, Amin J, Law M, Danta M, George J, Dore GJ (2017) Hepatocellular carcinoma risk following direct-acting antiviral HCV therapy: a systematic review, meta-analyses, and meta-regression. J Hepatol 67:1204–1212
Conti F, Buonfiglioli F, Scuteri A, Crespi C, Bolondi L, Caraceni P, Foschi FG, Lenzi M, Mazzella G, Verucchi G, Andreone P, Brillanti S (2016) Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. J Hepatol 65:727–733
Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, Hirose Y, Nagahashi M, Iso T, Fukutomi T, Ohishi M, Endo K, Uemura T, Nishito Y, Okuda S, Obata M, Kouno T, Imamura R, Tada Y, Obata R, Yasuda D, Takahashi K, Fujimura T, Pi J, Lee MS, Ueno T, Ohe T, Mashino T, Wakai T, Kojima H, Okabe T, Nagano T, Motohashi H, Waguri S, Soga T, Yamamoto M, Tanaka K, Komatsu M (2016) p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun 7:12030
Menegon S, Columbano A, Giordano S (2016) The Dual Roles of NRF2 in Cancer. Trends Mol Med 22:578–593
Acknowledgement
Lamp1 antibody was a kind gift from J.T. August (Johns Hopkins University). GFP-LC3 was a kind gift from Dr. Yoshimori (Osaka University).
Funding
This work was supported in part by JSPS KAKENHI Grant Number 18K07988, Grant-in-aid for Scientific Research (C) 22591052, the Research Program on Hepatitis from the Japan Agency for Medical Research and Development (AMED).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Honma, Y., Miyagawa, K., Hara, Y. et al. Correlation of hepatitis C virus-mediated endoplasmic reticulum stress with autophagic flux impairment and hepatocarcinogenesis. Med Mol Morphol 54, 108–121 (2021). https://doi.org/10.1007/s00795-020-00271-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00795-020-00271-5