Abstract
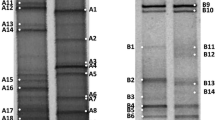
The prokaryotic communities of four salterns (Bingöl, Fadlum, Kemah, and Tuzlagözü) in Turkey were examined and compared using the cultivation and cultivation-independent methods [fluorescence in situ hybridization (FISH) and 454 pyrosequencing]. FISH analysis with universal probes revealed that feeding waters carried 1.6 × 102–1.7 × 103 cells mL−1, while crystallization ponds carried 3.8 × 106–2.0 × 107 cells mL−1 that were mostly haloarchaea, including square cells (except for Kemah). High-throughput 16S rRNA-based gene sequencing showed that the most frequent archaeal OTUs in Bingöl, Fadlum, Tuzlagözü, and Kemah samples were affiliated with Haloquadratum (76.8 %), Haloarcula (27.8 %), Halorubrum (49.6 %), and Halonotius (59.8 %), respectively. Bacteroidetes was the dominant bacterial phylum in Bingöl and Fadlum, representing 71.5 and 79.5 % of the bacterial OTUs (respectively), while the most abundant bacterial phylum found in the Kemah saltern was Proteobacteria (79.6 %). The majority of the bacterial OTUs recovered from Tuzlagözü belonged to the Cyanobacteria (35.7 %), Bacteroidetes (35.0 %), and Proteobacteria (25.5 %) phyla. Cultivation studies revealed that the archaeal isolates were closely related to the genera Halobacterium, Haloarcula, and Halorubrum. Bacterial isolates were confined to two phyla, Proteobacteria (Alphaproteobacteria and Gammaproteobacteria classes) and Bacteroidetes. Comparative analysis showed that members of the Euryarchaeota, Bacteroidetes, Proteobacteria, and Cyanobacteria phyla were major inhabitants of the solar salterns.









Similar content being viewed by others
References
Aktimur HT, Tekirli ME, Yurdakul ME (1990) Geology of the Sivas-Erzincan Tertiary basin. Bull Miner Res Explor Inst Turkey 111:21–30
Amann RI (1995) In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes. Molecular Microbial Ecology Manual. Kluwer academic Publishers, Dordrecht, pp 331–345
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Antón J, Llobet Brossa E, Rodríguez-Valera F, Amann R (1999) Fluorescence in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ Microbiol 1:517–523
Antón J, Rosselló-Mora R, Rodríguez-Valera F, Amann R (2000) Extremely halophilic bacteria in crystallizer ponds from solar salterns. Appl Environ Microbiol 66:3052–3057
Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R (2002) Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol 52:485–491
Baati H, Guermazi S, Amdouni R, Gharsallah N, Sghir A, Ammar E (2008) Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles 12:505–518
Baati H, Guermazi S, Gharsallah N, Sghir A, Ammar E (2010) Microbial community of salt crystals processed from Mediterranean seawater based on 16S rRNA analysis. Can J Microbiol 56:44–51
Benlloch S, López-López A, Casamayor EO, Øvreås L, Goddard V, Daae FL, Smerdon G, Massana R, Joint I, Thingstad F, Pedrós-Alió C, Rodríguez Valera F (2002) Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ Microbiol 4:349–360
Boujelben I, Gomariz M, Martínez-García M, Santos F, Peña A, López C, Antón J, Maalej S (2012) Spatial and seasonal prokaryotic community dynamics in ponds of increasing salinity of Sfax solar saltern in Tunisia. Antonie Van Leeuwenhoek 101:845–857
Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML (2004) Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl Environ Microbiol 70:5258–5265
Casamayor EO, Massana R, Benlloch S, Øvreås L, Díez B, Goddard VJ, Gasol JM, Joint I, Rodríguez-Valera F, Pedrós Alió C (2002) Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ Microbiol 4:338–348
Caton IR, Schneegurt MA (2012) Culture-independent analysis of the soil bacterial assemblage at the Great Salt Plains of Oklahoma. J Basic Microbiol 52:16–26
Caton TM, Caton IR, Witte LR, Schneegurt MA (2009) Archaeal diversity at the Great Salt Plains of Oklahoma described by cultivation and molecular analyses. Microb Ecol 58:519–528
Cifuentes A, Anton J, Benlloch S, Donnelly A, Herbert RA, Rodríguez-Valera F (2000) Prokaryotic diversity in Zostera noltii-colonized marine sediments. Appl Environ Microbiol 66:1715–1719
Çiner A, Koşun E (1996) Hafik Güneyindeki (Sivas Havzası) Oligo-Miyosen Yaşlı Çökellerin Stratigrafisi ve Sedimantolojisi. Turkish Assoc Pet Geol Bull 8:16–34
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2013) Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci 89:5685–5689
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469
Dillon J, Carlin M, Gutierrez A, Nguyen V, McLain N (2013) Patterns of microbial diversity along a salinity gradient in the Guerrero Negro solar saltern, Baja CA Sur, Mexico. Front Microbiol 4:399
Dyall-Smith M (2009) The halohandbook—protocols for haloarchaeal genetics, version 7.1. Martinsried, Germany, pp 12–15, 38–40. http://www.haloarchaea.com/resources/halohandbook/Halohandbook_2009_v7.1.pdf
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Fernández AB, León MJ, Vera B, Sánchez-Porro C, Ventosa A (2014) Metagenomic sequence of prokaryotic microbiota from an intermediate-salinity pond of a saltern in Isla Cristina, Spain. Genome Announc 2:e00045–e00114
Garcia-Pichel F, Nübel U, Muyzer G (1998) The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch Microbiol 169:469–482
Ghai R, Pašić L, Fernández AB, Martin-Cuadrado AB, Mizuno CM, McMahon KD, Papke RT, Stepanauskas R, Rodríguez-Brito B, Rohwer F, Sánchez-Porro C, Ventosa A, Rodríguez-Valera F (2011) New abundant microbial groups in aquatic hypersaline environments. Sci Rep 1:135
Gomariz M, Martínez-García M, Santos F, Rodriguez F, Capella-Gutiérrez S, Gabaldón T, Rosselló-Mora R, Meseguer I, Antón J (2015) From community approaches to single-cell genomics: the discovery of ubiquitous hyperhalophilic Bacteroidetes generalists. ISME J 9:16–31
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Harris JK, Caporaso JG, Walker JJ, Spear JR, Gold NJ, Robertson CE, Hugenholtz P, Goodrich J, McDonald D, Knights D, Marshall P, Tufo H, Knight R, Pace NR (2013) Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat. ISME J 7:50–60
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Jiang H, Dong H, Yu B, Liu X, Li Y, Ji S, Zhang CL (2007) Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan plateau. Environ Microbiol 9:2603–2621
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR (1985) Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci 82:6955–6959
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Makhdoumi-Kakhki A, Amoozegar MA, Kazemi B, Pašić L, Ventosa A (2012) Prokaryotic diversity in Aran-Bidgol salt lake, the largest hypersaline playa in Iran. Microbes Environ 27:87–93
Maturrano L, Santos F, Rosselló-Mora R, Antón J (2006) Microbial diversity in Maras salterns, a hypersaline environment in the Peruvian Andes. Appl Environ Microbiol 72:3887–3895
Mutlu MB, Martínez-García M, Santos F, Peña A, Guven K, Antón J (2008) Prokaryotic diversity in Tuz Lake, a hypersaline environment in Inland Turkey. FEMS Microbiol Ecol 65:474–483
Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ, Heidelberg KB, Banfield JF, Allen EE (2012) De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6:81–93
Nogales B, Moore ER, Abrham WR, Timmis KN (1999) Identification of the metabolically active members of a bacterial community in a polychlorinated biphenylpolluted moorland soil. Environ Microbiol 1:199–212
Oh D, Porter K, Russ B, Burns D, Dyall-Smith M (2010) Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles 14:161–169
Oren A (2002) Halophilic microorganisms and their environments. Kluwer Academic Publishers, Dordrecht, p 393
Oren A (2008) Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst 4:13
Oren A, Rodrı́guez-Valera F (2001) The contribution of halophilic Bacteria to the red coloration of saltern crystallizer ponds. FEMS Microbiol Ecol 36:123–130
Pašić L, Bartual SG, Ulrih NP, Grabnar M, Velikonja BH (2005) Diversity of halophilic Archaea in the crystallizers of an Adriatic solar saltern. FEMS Microbiol Ecol 54:491–498
Sabet S, Diallo L, Hays L, Jung W, Dillon JG (2009) Characterization of halophiles isolated from solar salterns in Baja California, Mexico. Extremophiles 13:643–656
Snaidr J, Amann R, Huber I, Ludwig W, Schleifer KH (1997) Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol 63:2884–2896
Sørensen KB, Canfield DE, Teske AP, Oren A (2005) Community composition of a hypersaline endoevaporitic microbial mat. Appl Environ Microbiol 71:7352–7365
Walsh DA, Papke RT, Doolittle WF (2005) Archaeal diversity along a soil salinity gradient prone to disturbance. Environ Microbiol 7:1655–1666
Wang YX, Liu JH, Xiao W, Ma XL, Lai YH, Li ZY, Ji KY, Wen ML, Cui XL (2013) Aliifodinibius roseus gen. nov., sp. nov., and Aliifodinibius sediminis sp. nov., two moderately halophilic bacteria isolated from salt mine samples. Int J Syst Evol Microbiol 63:2907–2913
Zhaxybayeva O, Stepanauskas R, Mohan NR, Papke RT (2013) Cell sorting analysis of geographically separated hypersaline environments. Extremophiles 17:265–275
Acknowledgments
We would like to thank Prof. Dr. Mike Dyall-Smith for his helpful and critical comments on this work. The authors would like to thank Dr. Dionysios Antonopoulos from the Argonne National Laboratory (USA) for the English language review. This work was supported by the Anadolu University Research Foundation [Grant Number 1109F153].
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Çınar, S., Mutlu, M.B. Comparative analysis of prokaryotic diversity in solar salterns in eastern Anatolia (Turkey). Extremophiles 20, 589–601 (2016). https://doi.org/10.1007/s00792-016-0845-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-016-0845-7