Abstract
GD-95 lipase from Geobacillus sp. strain 95 and its modified variants lacking N-terminal signal peptide and/or 10 or 20 C-terminal amino acids were successfully cloned, expressed and purified. To our knowledge, GD-95 lipase precursor (Pre-GD-95) is the first Geobacillus lipase possessing more than 80 % lipolytic activity at 5 °C. It has maximum activity at 55 °C and displays a broad pH activity range. GD-95 lipase was shown to hydrolyze p-NP dodecanoate, tricaprylin and canola oil better than other analyzed substrates. Structural and sequence alignments of bacterial lipases and GD-95 lipase revealed that the C-terminus forms an α helix, which is a conserved structure in lipases from Pseudomonas, Clostridium or Staphylococcus bacteria. This work demonstrates that 10 and 20 C-terminal amino acids of GD-95 lipase significantly affect stability and other physicochemical properties of this enzyme, which has never been reported before and can help create lipases with more specific properties for industrial application. GD-95 lipase and its modified variants GD-95-10 can be successfully applied to biofuel production, in leather and pulp industries, for the production of cosmetics or perfumes. These lipases are potential biocatalysts in processes, which require extreme conditions: low or high temperature, strongly acidic or alkaline environment and various organic solvents.



Similar content being viewed by others
References
Arpigny JL, Jaeger KE (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343(Pt 1):177–183
Balan A, Ibrahim D, Abdul Rahim R, Ahmad Rashid FA (2012) Purification and characterization of a thermostable lipase from Geobacillus thermodenitrificans IBRL-nra. Enzyme Res
Bornscheuer UT (2002) Microbial carboxyl esterases: classification, properties and applications in biocatalysis. FEMS Microbiol Rev 26:73–81
Carrasco-Lopez C, Godoy C, de las Rivas B, Fernandez-Lorente G, Palomo JM, Guisan JM, Fernandez-Lafuente R, Martinez-Ripoll M, Hermoso JA (2009) Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements. JBC 284(7):4365–4372
Charbonneau DM, Meddeb-Mouelhi F, Beauregard M (2010) A novel thermostable carboxylesterase from Geobacillus thermodenitrificans: evidence for a new carboxylesterase family. J Biochem 148(3):299–308
Cho AR, Yoo SK, Kim EJ (2000) Cloning, sequencing and expression in Escherichia coli of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol Lett 186:235–238
Dosztanyi Z, Fiser A, Simon I (1997) Stabilization centers in protein: identification, characterization and predictions. J Mol Biol 272:597–612
Dosztanyi Z, Magyar C, Tusnady GE, Simon I (2003) SCide: identification of stabilization centers in proteins. Bioinformatics 19:899–900
Ebrahimpour A, Rahman RNZRA, Basri M, Salleh AB (2011) High level expression and characterization of a novel thermostable, organic solvent tolerant, 1,3-regioselective lipase from Geobacillus sp. strain ARM. Bioresour Technol 102:6972–6981
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:29–37
Ghori MI, Iqbal MJ, Hameed A (2011) Characterization of a novel lipase from Bacillus sp. isolated from tannery wastes. Braz J Microbiol 42:22–29
Gomes N, Braga A, Teixeira JA, Belo I (2013) Impact of lipase-mediated hydrolysis of castor oil on γ-decalactone production by Yarrowia lipolytica. J Am Oil Chem Soc 90:1131–1137
Guncheva M, Zhiryakova D (2011) Catalytic properties and potential applications of Bacillus lipases. J Mol Catal B Enzym 68:1–21
Hasan F, Shah AA, Hameed A (2006) Industrial application of microbial lipases. Enzyme Microb Technol 39:235–251
Hasan F, Shah AA, Javed S, Hameed A (2010) Enzymes used in detergents: lipases. Afr J Biotechnol 9(31):4836–4844
Holm L, Rosenström P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38:545–549
Jaeger KE, Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13:390–397
Jaeger KE, Reetz MT (1998) Microbial lipases form versatile tolls for biotechnology. Tibtech 16:396–403
Jeong ST, Kim HK, Kim SJ, Chi SW, Pan JG, Oh TK, Ryu SE (2002) Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase. J Biol Chem 277(19):17041–17047
Jiang Y, Zhou X, Chen Z (2010) Cloning, expression, and biochemical characterization of a thermostable lipase from Geobacillus stearothermophilus JC. World J Microbiol Biotechnol 26:747–751
Katata L, Nagaraju V, Crouch AM (2006) Determination of ethylenediaminetetraacetic acid, ethylenediaminedisuccinic acid and iminodisuccinic acid in cosmetic products by capillary electrophoresis and high performance liquid chromatography. Anal Chim Acta 579:177–184
Kim HK, Park SY, Lee JK, Oh TK (1998) Gene cloning and characterization of thermostable lipase from Bacillus stearothermophilus L1. Biosci Biotechnol Biochem 62:66–71
Kuisiene N, Raugalas J, Stuknyte M, Chitavicius D (2007) Identification of the genus Geobacillus using genus-specific primers, based on the 16S-23S rRNA gene internal transcribed spacer. FEMS Microbiol Lett 277:165–172
Kwon DY, Rhee JS (1986) A simple and rapid colorimetric method for determination of free fatty acids for lipase assay. J Am Oil Chem Soc 63:89–92
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:380–685
Lee DW, Koh YS, Kim KJ, Kim BC, Choi HJ, Kim DS, Suhartono MT, Pyun YR (1999) Isolation and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol Lett 179:393–400
Leow TC, Rahman RNZRA, Basri M, Salleh AB (2004) High level expression of thermostable lipase from Geobacillus sp. strain T1. Biosci Biotechnol Biochem 68(1):96–103
Leow TC, Rahman RNZRA, Basri M, Salleh AB (2007) A thermoalkaliphilic lipase of Geobacillus sp. T1. Extremophiles 11:527–535
Levisson M, van der Oost J, Kengen SWM (2007) Characterization and structural modelling of a new type of thermostable esterase from Thermotoga maritime. FEBS J 274:2832–2842
Li H, Zhang X (2005) Characterization of thermostable lipase from thermophilic Geobacillus sp. TW1. Protein Expr Purif 42:153–159
Magyar C, Gromiha MM, Pujadas G, Tusnady GE, Simon I (2005) SRide: a server for identifying stabilizing residues in proteins. Nucleic Acids Res 33:303–305
Matsumura H, Yamamoto T, Leow TC, Mori T, Salleh AB, Basri M, Inoue T, Kai Y, Rahman RNZRA (2007) Novel cation-π interaction revealed by cristal structure of thermoalkalophilic lipase. Proteins 70:592–598
Nardini M, Dijkstra BW (1999) Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol 9(6):732–737
Olechnovič K, Kulberkytė E, Venclovas Č (2012) CAD-score: a new contact area difference-based function for evaluation of protein structural models. Proteins
Pei J, Kim BH, Grishin NV (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36(7):2295–2300
Petersen TN, Brunak S, Heijne G, Nielsen H (2011) SignalIP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Salleh AB, Rahim ASMA, Rahman RNZRA, Leow TC, Basri M (2012) The role of Arg157Ser in improving the compactness and stability of ARM lipase. J Comput Sci Syst Biol 5(2):039–046
Sambrook J, Rusell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Lab Press, New York
Sangeetha R, Arulpandi I, Geetha A (2011) Bacterial lipases as potential industrial biocatalysts: an overview. Res J Microbiol 6(1):1–24
Shariff FM, Rahman RNZRA, Basri M, Salleh AB (2011) A newly isolated thermostable lipase from Bacillus sp. Int J Mol Sci 12:2917–2934
Sharma D, Sharma B, Shukla AK (2011) Biotechnological approach of microbial lipase: a review. Biotechnology 10(1):23–40
Sinchaikul S, Sookkheo B, Phutrakul S, Pan FM, Chen ST (2001) Optimization of a thermostable lipase from Bacillus stearothermophilus P1: overexpression, purification, and characterization. Protein Expr Purif 22:388–398
Singh R, Gupta N, Goswami VK, Gupta R (2006) A simple activity staining protocol for lipases and esterases. Appl Microbiol Biotechnol 70:679–682
Tayyab M, Rashid N, Akhtar M (2011) Isolation and identification of lipase producing thermophilic Geobacillus sp. SBS-4S: cloning and characterization of the lipase. J Biosci Bioeng 111(3):272–278
Tyndall JDA, Sinchaikul S, Fothergill-Gilmore LA, Taylor P, Walkinshaw MD (2002) Crystal structure of a thermostable lipase from Bacillus stearothermophilus P1. J Mol Biol 323:859–869
Vehlow C, Stehr H, Winkelmann M, Duarte JM, Petzold L, Dinse J, Lappe M (2011) CMView: interactive contact map visualization and analysis. Bioinformatics 27(11):1573–1574
Vieille C, Zeikus GJ (2001) Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65(1):1–43
Villeneuve P, Muderhwa JM, Graille J, Haas MJ (2000) Customizing lipases for biocatalysis: a survey of chemical, physical and molecular biological approaches. J Mol Catal B Enzym 9:113–148
Wahab RA, Basri M, Rahman MBA, Rahman RNZRA, Salleh AB, Leow TC (2012a) Combination of oxyanion Gln114 mutation and medium engineering to influence the enantioselectivity of thermophilic lipase from Geobacillus zalihae. Int J Mol Sci 13:11666–11680
Wahab RA, Basri M, Rahman MBA, Rahman RNZRA, Salleh AB, Leow TC (2012b) Engineering catalytic efficience of thermophilic lipase from Geobacillus zalihae by hydrophobic residue mutation near the catalytic pocket. ABB 3:158–167
Wiederstein M, Sippl MJ (2007) ProSA-web:interactive web service for the recognition of errors in three-dimensional structures of protein. Nucleic Acids Res 35:407–410
Winkler UK, Stuckmann M (1979) Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J Bacteriol 138:663–670
Wu L, Liu B, Hong Y, Sheng D, Shen Y, Ni J (2010) Residue Tyr224 is critical for the thermostability of Geobacillus sp. RD-2 lipase. Biotechnol Lett 32:107–112
Yang Z, Zhang Y, Shen T, Xie Y, Mao Y, Ji C (2013) Cloning, expression and biochemical characterization of a novel, moderately thermostable GDSL family esterase from Geobacillus thermodenitrificans T2. J Biosci Bioeng 115(2):133–137
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformat 9:40
Zuo K, Zhang L, Yao H, Wang J (2010) Isolation and functional expression of a novel lipase gene isolated directly from oil-contaminated soil. Biochimica Polonica Acta 57(3):305–311
Acknowledgments
This work was supported by the MITA (Agency of Science, Innovation and Technology) program “Development of industrial biotechnology in Lithuania 2011-2013”, project “Innovative tools for cosmetic industry (COSMETIZYM)”, Grant No. MITA 31V-18.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Atomi.
Electronic supplementary material
Below is the link to the electronic supplementary material.
792_2013_605_MOESM4_ESM.tif
Supplementary material 4 (TIFF 1566 kb) 3D structure of GD-95 lipase. The structure predicted with I-TASSER server and image wasgenerated using Pymol (DeLano Scientific, Palo Alto, CA, USA). The arrows indicate beginning of N-and C -ends, α13 helix and catalytic amino acids of GD-95 lipase; 20 C-terminal amino acids are marked in black234x176mm (300 x 300 DPI)
792_2013_605_MOESM5_ESM.tif
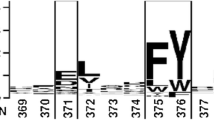
Supplementary material 5 (TIFF 3697 kb) Multiple alignment of 20 C-terminal amino acids from various bacterial lipases176x234mm (300 x 300 DPI)
792_2013_605_MOESM6_ESM.tif
Supplementary material 6 (TIFF 16860 kb) Effect of pH on the stability of GD-95 lipase and its modified variants. a: finely dotted line – Pre-GD-95; straight line - GD-95. b: finely dotted line – Pre-GD-95; straight line – Pre-GD-95-10; coarsely dottedline - GD- 95-10. The remaining activity was assayed under standard assay conditions after the purified recombinant lipases were pre-incubated at various pHs (1:4) at room temperature for 30 min. A specific activity of 270.0U/mg (GD-95), 100.7U/mg (Pre-GD-95), 113.0U/mg (GD-95-10), 32.3U/mg (Pre-GD-95-10) was recorded as 100%176x234mm (300 x 300 DPI)
792_2013_605_MOESM7_ESM.tif
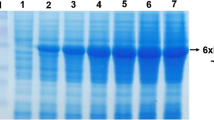
Supplementary material 7 (TIFF 1933 kb) SDS–PAGE (12%) analysis of recombinant Pre-GD-95 (a), GD-95-10 (c) and GD-95 (e) lipasespurified using affinity chromatography. M - PageRulerTM Unstained Protein Ladder. The arrows indicate thetarget proteins. Purification of Pre-GD-95 lipase (a): lane 1 - extracellular crude samples of enzyme; lane 2 -flow through; lane 3 - washing fraction; lanes 4–10 - 1-7 elution fraction, respectively. Purification of GD-95-10 lipase (c): lanes 1-5 - elution fraction; purification of GD-95 lipase (e): lanes 1-5 - elution fraction. band d - zymogram of purified Pre-GD-95 (b), GD-95 (in d Lane 1), Pre-GD-95-10 (in d Lane 3) and Pre-GD-95-20 (in d Lane 2) lipases. Zymogram results with GD-95-10 and GD-95-20 not shown234x176mm (300 x 300 DPI)
792_2013_605_MOESM8_ESM.tif
Supplementary material 8 (TIFF 1285 kb) Effect of pH on the enzyme activity of GD-95 lipase and its modified variants. a: finely dotted line –Pre-GD-95; straight line - GD-95. b: finely dotted line – Pre-GD-95; straight line – Pre-GD-95-10; coarselydotted line - GD-95-10. c: finely dotted line –Pre-GD-95; straight line – Pre-D-95-20; coarsely dotted line -GD-95-20. Enzyme activity was assayed under standard enzyme assay conditions. A specific activity of220.3U/mg (GD-95), 102.4U/mg (Pre-GD-95), 195.6U/mg (GD-95-10), 32.3U/mg (Pre-GD-95-10),2.7U/mg (Pre-GD-95-20), 5.1U/mg (GD-95-20) was recorded as 100% at pH 9176x234mm (300 x 300 DPI)
792_2013_605_MOESM9_ESM.tif
Supplementary material 9 (TIFF 16857 kb) Arrhenius plot for the determination of activation energy of Pre-GD-95 and GD-95 lipases. Thesquare marks Pre-GD-95, rhombs - GD-95 lipases. T is Kelvin temperature234x176mm (300 x 300 DPI)
Rights and permissions
About this article
Cite this article
Gudiukaitė, R., Gegeckas, A., Kazlauskas, D. et al. Influence of N- and/or C-terminal regions on activity, expression, characteristics and structure of lipase from Geobacillus sp. 95. Extremophiles 18, 131–145 (2014). https://doi.org/10.1007/s00792-013-0605-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-013-0605-x