Abstract
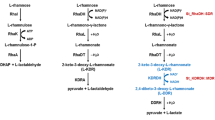
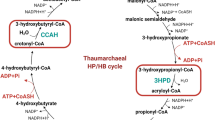
Aldehyde dehydrogenases (ALDHs) have been well established in all three domains of life and were shown to play essential roles, e.g., in intermediary metabolism and detoxification. In the genome of Sulfolobus solfataricus, five paralogs of the aldehyde dehydrogenases superfamily were identified, however, so far only the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) and α-ketoglutaric semialdehyde dehydrogenase (α-KGSADH) have been characterized. Detailed biochemical analyses of the remaining three ALDHs revealed the presence of two succinic semialdehyde dehydrogenase (SSADH) isoenzymes catalyzing the NAD(P)+-dependent oxidation of succinic semialdehyde. Whereas SSO1629 (SSADH-I) is specific for NAD+, SSO1842 (SSADH-II) exhibits dual cosubstrate specificity (NAD(P)+). Physiological significant activity for both SSO-SSADHs was only detected with succinic semialdehyde and α-ketoglutarate semialdehyde. Bioinformatic reconstructions suggest a major function of both enzymes in γ-aminobutyrate, polyamine as well as nitrogen metabolism and they might additionally also function in pentose metabolism. Phylogenetic studies indicated a close relationship of SSO-SSALDHs to GAPNs and also a convergent evolution with the SSADHs from E. coli. Furthermore, for SSO1218, methylmalonate semialdehyde dehydrogenase (MSDH) activity was demonstrated. The enzyme catalyzes the NAD+- and CoA-dependent oxidation of methylmalonate semialdehyde, malonate semialdehyde as well as propionaldehyde (PA). For MSDH, a major function in the degradation of branched chain amino acids is proposed which is supported by the high sequence homology with characterized MSDHs from bacteria. This is the first report of MSDH as well as SSADH isoenzymes in Archaea.






Similar content being viewed by others
Abbreviations
- ALDH:
-
Aldehyde dehydrogenase
- ED:
-
Entner–Doudoroff
- EMP:
-
Embden-Meyerhof-Parnas
- Sp:
-
Semi-phosphorylative
- Np:
-
Non-phosphorylative
- GA:
-
Glyceraldehyde
- α-KGSA:
-
α-Ketoglutarate semialdehyde
- α-KGSADH:
-
α-Ketoglutarate semialdehyde dehydrogenase
- GAP:
-
Glyceraldehyde-3-phosphate
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- GAPN:
-
Non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase
- MMSA:
-
Methylmalonate semialdehyde
- MSDH:
-
Methylmalonate semialdehyde dehydrogenase
- SSA:
-
Succinic semialdehyde
- SSADH:
-
Succinic semialdehyde dehydrogenase
- MSA:
-
Malonate semialdehyde
- PA:
-
Propionaldehyde
- GABA:
-
γ-Aminobutyric acid
References
Ahmed H, Ettema TJG, Tjaden B, Geerling ACM, Van Der Oost J, Siebers B (2005) The semi-phosphorylative Entner–Doudoroff pathway in hyperthermophilic archaea: a re-evaluation. Biochem J 390:529–540
Bannerjee D, Sanders LE, Sokatch JR (1970) Properties of purified methylmalonate semialdehyde dehydrogenase of Pseudomonas aeruginosa. J Biol Chem 245:1828–1835
Bchini R, Dubourg-Gerecke H, Rahuel-Clermont S, Aubry A, Branlant G, Didierjean C, Talfournier F (2012) Adenine binding mode is a key factor in triggering the early release of NADH in coenzyme A-dependent methylmalonate semialdehyde dehydrogenase. J Biol Chem 287:31095–31103
Brouns SJJ, Walther J, Snijders APL, Van De Werken HJG, Willemen HLDM, Worm P, De Vos MGJ, Andersson A, Lundgren M, Mazon HFM, Van Den Heuvel RHH, Nilsson P, Salmon L, De Vos WM, Wright PC, Bernander R, Van Der Oost J (2006) Identification of the missing links in prokaryotic pentose oxidation pathways: evidence for enzyme recruitment. J Biol Chem 281:27378–27388
Brunner N, Siebers B, Hensel R (2001) Role of two different glyceraldehyde-3-phosphate dehydrogenases in controlling the reversible Embden-Meyerhof-Parnas pathway in Thermoproteus tenax: regulation on protein and transcript level. Extremophiles 5:101–109
Cacciapuoti G, Porcelli M, Carteni-Farina M (1986) Purification and characterization of propylamine transferase from Sulfolobus solfataricus, an extreme thermophilic archaebacterium. Eur J Biochem 161:263–271
Cash C, Ciesielski L, Maitre M, Mandel P (1977) Purification and properties of rat brain succinic semialdehyde dehydrogenase. Biochimie 59:257–268
Cash CD, Maitre M, Ossola L, Mandel P (1978) Purification and properties of two succinate semialdehyde dehydrogenases from human brain. Biochim Biophys Acta 524:26–36
Crestia D, Guerard C, Bolte J, Demuynck C (2001) Rabbit muscle aldolase (RAMA) as a catalyst in a new approach for the synthesis of 3-deoxy-d-manno-2-octulosonic acid and analogues. J Mol Catal B Enzym 11:207–212
Donnelly MI, Cooper RA (1981) Two succinic semialdehyde dehydrogenases are induced when Escherichia coli K-12 is grown on γ-aminobutyrate. J Bacteriol 145:1425–1427
Ettema TJG, Ahmed H, Geerling ACM, Van Der Oost J, Siebers B (2008) The non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) of Sulfolobus solfataricus: a key-enzyme of the semi-phosphorylative branch of the Entner- Doudoroff pathway. Extremophiles 12:75–88
Fuhrer T, Chen L, Sauer U, Vitkup D (2007) Computational prediction and experimental verification of the gene encoding the NAD+/NADP+-dependent succinate semialdehyde dehydrogenase in Escherichia coli. J Bacteriol 189:8073–8078
Grochowski LL, Xu H, White RH (2006) Identification of lactaldehyde dehydrogenase in Methanocaldococcus jannaschii and its involvement in production of lactate for F420 biosynthesis. J Bacteriol 188:2836–2844
Grogan DW (1989) Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J Bacteriol 171:6710–6719
Kang JH, Park YB, Huh T-L, Lee W-H, Choi M-S, Kwon O-S (2005) High-level expression and characterization of the recombinant enzyme, and tissue distribution of human succinic semialdehyde dehydrogenase. Protein Expr Purif 44:16–22
Kardinahl S, Schmidt CL, Hansen T, Anemüller S, Petersen A, Schäfer G (1999) The strict molybdate-dependence of glucose-degradation by the thermoacidophile Sulfolobus acidocaldarius reveals the first crenarchaeotic molybdenum containing enzyme—an aldehyde oxidoreductase. Eur J Biochem 260:540–548
Kedishvili NY, Goodwin GW, Popov KM, Harris RA (2000) Mammalian methylmalonate-semialdehyde dehydrogenase. Methods Enzymol 324:207–218
Kim S, Lee SB (2005) Identification and characterization of Sulfolobus solfataricus d-gluconate dehydratase: a key enzyme in the non-phosphorylated Entner–Doudoroff pathway. Biochem J 387:271–280
Kim S, Lee SB (2006) Characterization of Sulfolobus solfataricus 2-keto-3-deoxy-d-gluconate kinase in the modified Entner–Doudoroff pathway. Biosci Biotechnol Biochem 70:1308–1316
Kockelkorn D, Fuchs G (2009) Malonic semialdehyde reductase, succinic semialdehyde reductase, and succinyl-coenzyme A reductase from Metallosphaera sedula: enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in Sulfolobales. J Bacteriol 191:6352–6362
Koo PH, Adams E (1974) α Ketoglutaric semialdehyde dehydrogenase of Pseudomonas. Properties of the separately induced isoenzymes. J Biol Chem 249:1704–1716
Kraus GA, Kevin L (1984) A direct synthesis of ω-bromo-1-alkenes. Synthesis 10:885
Kupiecki FP, Coon MJ (1960) Methylmalonic semialdehyde. Biochem Prep 7:69–71
Lamble HJ, Heyer NI, Bull SD, Hough DW, Danson MJ (2003) Metabolic pathway promiscuity in the archaeon Sulfolobus solfataricus revealed by studies on glucose dehydrogenase and 2-keto-3-deoxygluconate aldolase. J Biol Chem 278:34066–34072
Lamble HJ, Theodossis A, Milburn CC, Taylor GL, Bull SD, Hough DW, Danson MJ (2005) Promiscuity in the part-phosphorylative Entner–Doudoroff pathway of the archaeon Sulfolobus solfataricus. FEBS Lett 579:6865–6869
Lei Y, Pawelek PD, Powlowski J (2008) A shared binding site for NAD+ and coenzyme A in an acetaldehyde dehydrogenase involved in bacterial degradation of aromatic compounds. Biochemistry 47:6870–6882
Lindahl R (1992) Aldehyde dehydrogenases and their role in carcinogenesis. Crit Rev Biochem Mol Biol 27:283–335
Ludewig F, Hüser A, Fromm H, Beauclair L, Bouche N (2008) Mutants of GABA transaminase (POP2) suppress the severe phenotype of succinic semialdehyde dehydrogenase (ssadh) mutants in arabidopsis. PLoS One 3(10):e3383
Lütke-Eversloh T, Steinbüchel A (1999) Biochemical and molecular characterization of a succinate semialdehyde dehydrogenase involved in the catabolism of 4-hydroxybutyric acid in Ralstonia eutropha. FEMS Microbiol Lett 181:63–71
Macritchie JA, Silcock A, Willis CL (1997) Enantioselective synthesis of unsaturated α-hydroxy acids. Tetrahedron Asymmetry 8:3895–3902
Massey LK, Sokatch JR, Conrad RS (1976) Branched chain amino acid catabolism in bacteria. Bacteriol Rev 40:42–54
Milburn CC, Lamble HJ, Theodossis A, Bull SD, Hough DW, Danson MJ, Taylor GL (2006) The structural basis of substrate promiscuity in glucose dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus. J Biol Chem 281:14796–14804
Nunn CEM, Johnsen U, Schönheit P, Fuhrer T, Sauer U, Hough DW, Danson MJ (2010) Metabolism of pentose sugars in the hyperthermophilic archaea Sulfolobus solfataricus and Sulfolobus acidocaldarius. J Biol Chem 285:33701–33709
Oost JVD, Siebers B (2007) The glycolytic pathways of archaea: evolution by tinkering. Blackwell Publishing Ltd, New York
Reher M, Schönheit P (2006) Glyceraldehyde dehydrogenases from the thermoacidophilic euryarchaeota Picrophilus torridus and Thermoplasma acidophilum, key enzymes of the non-phosphorylative Entner–Doudoroff pathway, constitute a novel enzyme family within the aldehyde dehydrogenase superfamily. FEBS Lett 580:1198–1204
Rosemblatt MS, Callewaert DM, Tchen TT (1973) Succinic semialdehyde dehydrogenase from a Pseudomonas species. II. Physical and immunochemical properties of the enzyme. J Biol Chem 248:6014–6018
Rothacker B, Werr M, Ilg T (2008) Succinic semialdehyde dehydrogenase from the parasitic cattle tick Rhipicephalus microplus: gene identification, biochemical characterization and comparison with the mouse ortholog. Mol Biochem Parasitol 161:32–43
Sánchez LB (1998) Aldehyde dehydrogenase (CoA-acetylating) and the mechanism of ethanol formation in the amitochondriate protist, Giardia lamblia. Arch Biochem Biophys 354:57–64
Sànchez M, Alvarez MA, Balaña R, Garrido-Pertierra A (1988) Properties and functions of two succinic-semialdehyde dehydrogenases from Pseudomonas putida. Biochimica et Biophysica Acta (BBA)/Protein Struct Mol 953:249–257
Sànchez M, Fernandez J, Martin M, Gibello A, Garrido-Pertierra A (1989) Purification and properties of two succinic semialdehyde dehydrogenases from Klebsiella pneumoniae. Biochimica et Biophysica Acta—Gen Subj 990:225–231
She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CCY, Clausen IG, Curtis BA, De Moors A, Erauso G, Fletcher C, Gordon PMK, Heikamp-de Jong I, Jeffries AC, Kozera CJ, Medina N, Peng X, Thi-Ngoc HP, Redder P, Schenk ME, Theriault C, Tolstrup N, Charlebois RL, Doolittle WF, Duguet M, Gaasterland T, Garrett RA, Ragan MA, Sensen CW, Van Der Oostg J (2001) The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA 98:7835–7840
Snijders APL, Walther J, Peter S, Kinnman I, De Vos MGJ, Van De Werken HJG, Brouns SJJ, Van Der Oost J, Wright PC (2006) Reconstruction of central carbon metabolism in Sulfolobus solfataricus using a two-dimensional gel electrophoresis map, stable isotope labelling and DNA microarray analysis. Proteomics 6:1518–1529
Sohling B, Gottschalk G (1993) Purification and characterization of a coenzyme-A-dependent succinate-semialdehyde dehydrogenase from Clostridium kluyveri. Eur J Biochem 212:121–127
Sophos NA, Vasiliou V (2003) Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem Biol Interact 143–144:5–22
Sreerama L, Sladek NE (1993) Identification and characterization of a novel class 3 aldehyde dehydrogenase overexpressed in a human breast adenocarcinoma cell line exhibiting oxazaphosphorine-specific acquired resistance. Biochem Pharmacol 45:2487–2505
Stines-Chaumeil C, Talfournier F, Branlant G (2006) Mechanistic characterization of the MSDH (methylmalonate semialdehyde dehydrogenase) from Bacillus subtilis. Biochem J 395:107–115
Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, Von Mering C (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39:D561–D568
Talfournier F, Stines-Chaumeil C, Branlant G (2011) Methylmalonate semialdehyde dehydrogenase from Bacillus subtilis: substrate specificity and coenzyme A binding. J Biol Chem 286(25):21971–21981
Tamaki N, Nakamura M, Kimura K, Hama T (1977) Purification and properties of aldehyde dehydrogenase from Saccharomyces cerevisiae. J Biochem 82:73–79
Tokunaga M, Nakano Y, Kitaoka S (1976) Separation and properties of the NAD linked and NADP linked isozymes of succinic semialdehyde dehydrogenase in Euglena gracilis z. Biochim Biophys Acta 429:55–62
Van de Casteele M, Demarez M, Legrain C, Glansdorff N, Pierard A (1990) Pathways of arginine biosynthesis in extreme thermophilic archaeo- and eubacteria. J Gen Microbiol 136:1177–1183
Van Der Does C, Den Blaauwen T, De Wit JG, Manting EH, Groot NA, Fekkes P, Driessen AJM (1996) SecA is an intrinsic subunit of the Escherichia coli preprotein translocase and exposes its carboxyl terminus to the periplasm. Mol Microbiol 22:619–629
Vasiliou V, Pappa A, Petersen DR (2000) Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem Biol Interact 129:1–19
Wagner M, Berkner S, Ajon M, Driessen AJM, Lipps G, Albers SV (2009) Expanding and understanding the genetic toolbox of the hyperthermophilic genus Sulfolobus. Biochem Soc Trans 37:97–101
Zaparty M, Zaigler A, Stamme C, Soppa J, Hensel R, Siebers B (2008) DNA microarray analysis of central carbohydrate metabolism: glycolytic/gluconeogenic carbon switch in the hyperthermophilic crenarchaeum Thermoproteus tenax. J Bacteriol 190:2231–2238
Zaparty M, Esser D, Gertig S, Haferkamp P, Kouril T, Manica A, Pham TK, Reimann J, Schreiber K, Sierocinski P, Teichmann D, van Wolferen M, von Jan M, Wieloch P, Albers SV, Driessen AJM, Klenk HP, Schleper C, Schomburg D, van der Oost J, Wright PC, Siebers B (2009) “Hot standards” for the thermoacidophilic archaeon Sulfolobus solfataricus. Extremophiles 14:119–142
Zillig W, Stetter KO, Wunderl S (1980) The Sulfolobus-’Caldariella’ group: taxonomy on the basis of the structure of DNA-dependent RNA polymerases. Arch Microbiol 125:259–269
Acknowledgments
The project was partially performed in the course of the Sulfolobus Systems Biology “SulfoSYS” (SysMO, BMBF) and the “Hot signal transduction in Sulfolobus” (DFG) project. DE was supported by the respective grants from the BMBF (0315004A) and DFG (SI 642/10-1).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Albers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Esser, D., Kouril, T., Talfournier, F. et al. Unraveling the function of paralogs of the aldehyde dehydrogenase super family from Sulfolobus solfataricus . Extremophiles 17, 205–216 (2013). https://doi.org/10.1007/s00792-012-0507-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-012-0507-3