Abstract
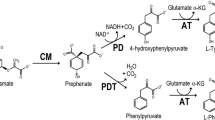
In the aromatic amino acid biosynthesis pathway, chorismate presents a branch point intermediate that is converted to tryptophan, phenylalanine (Phe), and tyrosine (Tyr). In bacteria, three enzymes catalyze the conversion of chorismate to hydroxyphenylpyruvate or pyruvate. The enzymes, chorismate mutase (CM), prephenate dehydratase (PDT), and prephenate dehydrogenase (PDHG) are either present as distinct proteins or fusions combining two activities. Gene locus AF0227 of Archaeoglobus fulgidus is predicted to encode a fusion protein, AroQ, containing all three enzymatic domains. This work describes the first characterization of a trifunctional AroQ. The A. fulgidus aroQ gene was cloned and overexpressed in Escherichia coli. The recombinant protein purified as a homohexamer with specific activities of 10, 0.51, and 50 U/mg for CM, PDT, and PDHG, respectively. Tyr at 0.5 mM concentration inhibited PDHG activity by 50%, while at 1 mM PDT was activated by 70%. Phe at 5 μM inhibited PDT activity by 66% without affecting the activity of PDHG. A fusion of CM, PDT, and PDHG domains is evident in the genome of only one other organism sequenced to date, that of the hyperthermophilic archaeon, Nanoarchaeum equitans. Such fusions of contiguous activities in a biosynthetic pathway may constitute a primitive strategy for the efficient processing of labile metabolites.





Similar content being viewed by others
References
Bonvin J, Aponte RA, Marcantonio M, Singh S, Christendat D, Turnbull JL (2006) Biochemical characterization of prephenate dehydrogenase form the hyperthermophilic bacterium Aquifex aeolicus. Protein Sci 15:1417–1432
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890
Cotton RG, Gibson F (1965) The biosynthesis of phenylalanine and tyrosine; enzymes converting chorismic acid into prephenic acid and their relationships to prephenate dehydratase and prephenate dehydrogenase. Biochim Biophys Acta 100:76–88
D’Amico S, Marx J-C, Gerday C, Feller G (2003) Activity–stability relationships in extremophilic enzymes. J Biol Chem 278:7891–7896
Davidson BE, Blackburn EH, Dopheide TAA (1972) Chorismate mutase-prephenate dehydratase from Escherichia coli K-12. J Biol Chem 247:4441–4446
Dawson RMC, Elliott DC, Elliott WH, Jones KM (1989) Data for biochemical research, 3rd edn. Oxford University Press, New York
Dosselaere F, Vanderleyden J (2001) A metabolic node in action: Chorimate-utilizing enzymes in microorganisms. Crit Rev Microbiol 27:75–131
Euverink GJW, Wolters DJ, Dijkhuizen L (1995) Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of wild-type and deregulated mutant protein. Biochem J 308:313–320
Felsenstein J (1993) PHYLIP (Phylogeny Inference Package), version 3.5c, distributed by the author. Department of Genetics, University of Washington, Seattle
Fischer RS, Bonner CA, Boone DR, Jensen RA (1993) Clues from a halophilic methanogen about aromatic amino acid biosynthesis in archaebacteria. Arch Microbiol 160:440–446
Friedrich CG, Friedrich B, Schlegel HG (1976) Regulation of chorismate mutase-prephenate dehydratase and prephenate dehydrogenase from Alcaligenes eutrophus. J Bacteriol 126:723–732
Helmstaedt K, Heinrich G, Merkl R, Braus GH (2004) Chorismate mutase of Thermus thermophilus is a monofunctional AroH class enzyme inhibited by tyrosine. Arch Microbiol 181:195–203
Heyde E, Morrison JF (1978) Kinetic studies on the reactions catalyzed by chorismate mutase-prephenate dehydrogenase from Aerobacter aerogenes. Biochemistry 17:1573–1580
Kim SK et al (2006) Biochemical and structural characterization of the secreted chorismate mutase (Rv1885c) from Mycobacterium tuberculosis H37Rv: an *AroQ enzyme not regulated by the aromatic amino acids. J Bacteriol 188:8638–8648
Kleeb AC, Kast P, Hilvert D (2006) A monofunctional and thermostable prephenate dehydratase from the archaeon Methanocaldococcus jannaschii. Biochemistry 45:14101–14110
Koch GLE, Shaw DC, Gibson F (1970) Tyrosine biosynthesis in Aerobacter aerogenes. Purification and properties of chorismate mutase-prephenate dehydrogenase. Biochim Biophys Acta 212:375–386
Koch GLE, Shaw DC, Gibson F (1971) The purification and characterization of chorismate mutase-prephenate dehydrogenase from Escherichia coli K12. Biochim Biophys Acta 229:795–804
Lim S, Schröder I, Monbouquette HG (2004) A thermostable shikimate 5-dehydrogenase from the archaeon Archaeoglobus fulgidus. FEMS Microbiol Lett 238:101–106
MacBeath G, Kast P, Hilvert D (1998) A small, thermostable, and monofunctional chorismate mutase from the archaeon Methanococcus jannaschii. Biochemistry 37:10062–10073
Perbal B (1988) A practical guide to molecular cloning, 2nd edn. Wiley, New York
Porat I, Waters BW, Teng Q, Whitman WB (2004) Two biosynthetic pathways for aromatic amino acids in the archaeon Methanococcus maripaludis. J Bacteriol 186:4940–4950
Riepl RG, Glover GI (1978) Purification of prephenate dehydratase from Bacillus subtilis. Arch Biochem Biophys 191:192–197
Sasso S, Ramakrishnan C, Gamper M, Hilvert D, Kast P (2005) Characterization of the secreted chorismate mutase from the pathogen Mycobacterium tuberculosis. FEBS J 272:375–389
Schmit JC, Zalkin H (1971) Chorismate mutase-prephenate dehydratase. Phenylalanine-induced dimerization and its relationship to feedback inhibition. J Biol Chem 246:6002–6010
Schröder I, Vadas A, Johnson E, Lim S, Monbouquette HG (2004) A novel archaeal alanine dehydrogenase homologous to ornithine cyclodeaminase and mu-crystallin. J Bacteriol 186:7680–7689
Waters E et al (2003) The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci U S A 100:12984–12988
Xia T, Song J, Zhao G, Aldrich H, Jensen RA (1993) The aroQ-encoded monofunctional chorismate mutase (CM-F) protein is a periplasmic enzyme in Erwinia herbicola. J Bacteriol 175:4729–4737
Yanai I, Wolf YI, Koonin EV (2002) Evolution of gene fusions: horizontal transfer versus independent events. Genome Biol 3:1–13
Zhang S, Pohner G, Kongsaeree P, Wilson DB, Clardy J, Ganem B (1998) Chorismate mutase-prephenate dehydratase from Escherichia coli. Study of catalytic and regulatory domains using genetically engineered proteins. J Biol Chem 273:6248–6253
Acknowledgments
We thank Chi-Hee Kim for the construction of plasmid pIS85. This work was supported by the National Science Foundation (BES-9911718) and the US Department of Commerce/NIST (Cooperative Agreement 70NANBOH0064).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Huang.
Rights and permissions
About this article
Cite this article
Lim, S., Springstead, J.R., Yu, M. et al. Characterization of a key trifunctional enzyme for aromatic amino acid biosynthesis in Archaeoglobus fulgidus . Extremophiles 13, 191–198 (2009). https://doi.org/10.1007/s00792-008-0209-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-008-0209-z