Abstract
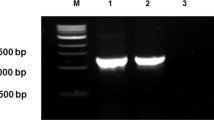
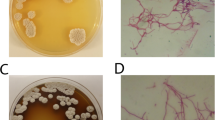
The isolation and phylogenetic characterization of acidophilic moderate thermophilic bacteria from different locations of uranium mines and a uranium processing mill in Pakistan is reported. The dominant culturable bacteria found were related to Sulfobacillus thermosulfidooxidans in all the samples analyzed. Different strains displayed different levels of identity (95–97%) to16S rDNA of known strains of this species, indicating group heterogeneity. Genomic DNA from five isolates was subjected to amplification using integron-specific primers HS286 and HS287. Recovery of different integron-linked genes from one of the isolates indicated the usefulness of this approach for gene mining in place of traditional gene recovery methodologies.


Similar content being viewed by others
References
Bennett PM (1999) Integrons and gene cassettes: a genetic construction kit for bacteria. J Antimicrob Chemother 43:1–4
Berthelot D, Leduc LG, Ferroni GD (1993) Temperature studies of iron-oxidizing autotrophs and acidophilic heterotrophs isolated from uranium mines. Can J Microbiol 39:384–388
Collis CM, Hall RM (1992) Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol Microbiol 6:2875–2885
Hall RM, Collis CM (1995) Mobile gene cassettes and integrons: capture and spread of genes by site specific recombination. Mol Microbiol 15:593–600
Hall RM, Brookes DE, Stokes HW (1991) Site-specific insertion of genes into integrons: role of the 59 base-element and determination of the recombination cross-over point. Mol Microbiol 5:1941–1959
Johnson DB (1995) Selective solid media for isolating and enumerating acidophilic bacteria. J Microbiol Methods 23:205–218
Johnson DB (1998) Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol Ecol 27:307–317
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic, New York, pp 21–132
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–148
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Norris PR, Johnson DB (1998) Acidophilic microorganisms. In: Horikoshi K, Grant WD (eds) Extremophiles: microbial life in extreme environments. Wiley-Liss, New York, pp 133–153
Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304
Rowe-Magnus DA, Mazel D (2001) Integrons: natural tools for bacterial genome evolution. Curr Opin Microbiol 4:565–569
Rowe-Magnus DA, Guerot AM, Ploncard P, Dychinco B, Davies J, Mazel D (2001) The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc Natl Acad Sci U S A 98:652–657
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Smalla K, Krogerrecklenfort E, Heuer H, Dejonghe W, Top E, Osborn M, Niewint J, Tebbe C, Barr M, Bailey M, Gretaed A, Thomas C, Turner S, Young P, Nikolakopoulou D, Karagouni A, Wolters A, van Elsas JD, Dronen K, Sandaa R, Borin S, Brabhu J, Grohmann E, Sobecky P (2000) PCR-based detection of mobile genetic elements in total community DNA. Microbiology 146:1256–1257
Stokes HW, Holmes AJ, Nield BS, Holley MP, Nevalainen KMH, Mabbutt BC, Gillings MR (2001) Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl Environ Microbiol 67:5240–5246
Van de Peer Y, De Wachter R (1993) TREECON: a software package for the construction and drawing of evolutionary trees. Comput Appl Biosci 9:177–182
Acknowledgments
M. Afzal Ghauri greatly acknowledges the provision of a Postdoctoral Fellowship by the Ministry of Science and Technology, Government of Pakistan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Horikoshi
Rights and permissions
About this article
Cite this article
Ghauri, M.A., Khalid, A.M., Grant, S. et al. Phylogenetic analysis of different isolates of Sulfobacillus spp. isolated from uranium-rich environments and recovery of genes using integron-specific primers. Extremophiles 7, 341–345 (2003). https://doi.org/10.1007/s00792-003-0354-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-003-0354-3