Abstract
Objectives
To investigate the variant of an amelogenesis imperfecta (AI) family and to explore the function of the FAM83H (family with sequence similarity 83 member H) in the enamel formation.
Materials and methods
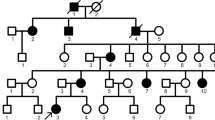
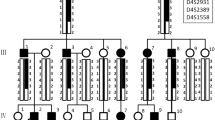
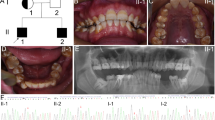
We investigated a five-generation Chinese family diagnosed with AI; clinical data was collected, whole-exome sequencing (WES) was conducted to explore the pathogenic gene and variants and Sanger sequencing was used to verify the variants. The three-dimensional protein structures of wild-type and mutant FAM83H were predicted using alpha fold 2. To study the possible regulatory function of Fam83h on amelogenesis, immunolocalization was performed to observe the expression of Fam83h protein in Sprague–Dawley rat postnatal incisors. The mRNA and protein level of amelogenin, enamelin, kallikrein-related peptidase-4 and ameloblastin were also detected after the Fam83h was knocked down by small interfering RNA (siRNA) in HAT-7 cells.
Results
A known nonsense variant (c.973 C > T) in exon 5 of FAM83H gene was found in this family, causing a truncated protein (p.R325X). Immunolocalization of Fam83h in Sprague–Dawley rat postnatal incisors showed that Fam83h protein expression was detected in presecretory and secretory stages. When Fam83h expression was reduced by siRNA, the expression of amelogenin, enamelin, kallikrein-related peptidase-4 decreased. However, the expression of ameloblastin increased.
Conclusions
FAM83H gene variant (c.973 C > T) causes AI. FAM83H regulates the secretion of enamel matrix proteins and affects ameloblast differentiation.
Clinical relevance
This study provided that FAM83H variants could influence enamel formation and provided new insights into the pathogenesis of AI.




Similar content being viewed by others
References
Bartlett JD (2013) Dental enamel development: proteinases and their enamel matrix substrates. ISRN Dent 2013:684607. https://doi.org/10.1155/2013/684607
Lacruz RS, Habelitz S, Wright JT, Paine ML (2017) Dental enamel formation and inplications for oral health and disease. Physiol Rev 97(3):939–993. https://doi.org/10.1152/physrev.00030.2016
Sabandal MM, Schäfer E (2016) Amelogenesis imperfecta: review of diagnostic findings and treatment concepts. Odontology 104(3):245–256. https://doi.org/10.1007/s10266-016-0266-1
Simmer JP, Papagerakis P, Smith CE, Fisher DC, Rountrey AN, Zheng L, Hu JCC (2010) Regulation of dental enamel shape and hardness. J Dent Res 89(10):1024–1038. https://doi.org/10.1177/0022034510375829
Smith CEL, Poulter JA, Antanaviciute A, Kirkham J, Brookes SJ, Inglehearn CF, Mighell AJ (2017) Amelogenesis imperfecta; genes, proteins and pathways. Front Physiol 8:435. https://doi.org/10.3389/fphys.2017.00435
Stephanopoulos G, Garefalaki ME, Lyroudia K (2005) Genes and related proteins involved in amelogenesis imperfecta. J Dent Res 84(12):1117–1126. https://doi.org/10.1177/154405910508401206
Lu Y, Papagerakis P, Yamakoshi Y, Hu JCC, Bartlett JD, Simmer JP (2008) Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem 389(6):695–700. https://doi.org/10.1515/BC.2008.080
Crawford PJM, Aldred M, Bloch-Zupan A (2007) Amelogenesis imperfecta. Orphanet J Rare Dis 2:17. https://doi.org/10.1186/1750-1172-2-17
Lee SK, Lee KE, Jeong TS, Hwang YH, Kim S, Hu JC, Simmer JP, Kim JW (2011) FAM83H mutations cause ADHCAI and alter intracellular protein localization. J Dent Res 90(3):377–381. https://doi.org/10.1177/0022034510389177
Zhang C, Song Y, Bian Z (2015) Ultrastructural analysis of the teeth affected by amelogenesis imperfecta resulting from FAM83H mutations and review of the literature. Oral Surgery Oral Medicine Oral Pathology Oral Radiology 119(2):E69–E76. https://doi.org/10.1016/j.oooo.2014.09.002
Hu JCC, Chun Y-HP, Al Hazzazzi T, Simmer JP (2007) Enamel formation and amelogenesis imperfecta. Cells Tissues Organs 186(1):78–85. https://doi.org/10.1159/000102683
Hyun HK, Lee SK, Lee KE, Kang HY, Kim EJ, Choung PH, Kim JW (2009) Identification of a novel FAM83H mutation and microhardness of an affected molar in autosomal dominant hypocalcified amelogenesis imperfecta. Int Endod J 42(11):1039–1043. https://doi.org/10.1111/j.1365-2591.2009.01617.x
Smith CEL, Whitehouse LLE, Poulter JA, Wilkinson Hewitt L, Nadat F, Jackson BR, Manfield IW, Edwards TA, Rodd HD, Inglehearn CF et al (2020) A missense variant in specificity protein 6 (SP6) is associated with amelogenesis imperfecta. Hum Mol Genet 29(9):1417–1425. https://doi.org/10.1093/hmg/ddaa041
Zheng Y, Lu T, Chen J, Li M, Xiong J, He F, Gan Z, Guo Y, Zhang L, Xiong F (2021) The gain-of-function FAM83H mutation caused hypocalcification amelogenesis imperfecta in a Chinese family. Clin Oral Investig 25(5):2915–2923. https://doi.org/10.1007/s00784-020-03609-6
Wang SK, Zhang H, Hu CY, Liu JF, Chadha S, Kim JW, Simmer JP, Hu JCC (2021) FAM83H and autosomal dominant hypocalcified amelogenesis imperfecta. J Dent Res 100(3):293–301. https://doi.org/10.1177/0022034520962731
Sriwattanapong K, Nitayavardhana I, Theerapanon T, Thaweesapphithak S, Chantarawaratit PO, Garuyakich R, Phokaew C, Porntaveetus T, Shotelersuk V (2021) Age-related dental phenotypes and tooth characteristics of FAM83H-associated hypocalcified amelogenesis imperfecta. Oral Dis. https://doi.org/10.1111/odi.13780
Kantaputra PN, Intachai W, Auychai P (2016) All enamel is not created equal: supports from a novel FAM83H mutation. Am J Med Genet A 170(1):273–276. https://doi.org/10.1002/ajmg.a.37406
Song YL, Wang CN, Zhang CZ, Yang K, Bian Z (2012) Molecular characterization of amelogenesis imperfecta in Chinese patients. Cells Tissues Organs 196(3):271–279. https://doi.org/10.1159/000334210
Wright JT, Torain M, Long K, Seow K, Crawford P, Aldred MJ, Hart PS, Hart TC (2011) Amelogenesis imperfecta: genotype-phenotype studies in 71 families. Cells Tissues Organs 194(2–4):279–283. https://doi.org/10.1159/000324339
Haubek D, Gjørup H, Jensen LG, Juncker I, Nyegaard M, Børglum AD, Poulsen S, Hertz JM (2011) Limited phenotypic variation of hypocalcified amelogenesis imperfecta in a Danish five-generation family with a novel FAM83H nonsense mutation. Int J Paediatr Dent 21(6):407–412. https://doi.org/10.1111/j.1365-263X.2011.01142.x
Chan HC, Estrella NM, Milkovich RN, Kim JW, Simmer JP, Hu JC (2011) Target gene analyses of 39 amelogenesis imperfecta kindreds. Eur J Oral Sci 119(Suppl 1):311–323. https://doi.org/10.1111/j.1600-0722.2011.00857.x
Ding Y, Estrella MRP, Hu YY, Chan HL, Zhang HD, Kim JW, Simmer JP, Hu JCC (2009) Fam83h is associated with intracellular vesicles and ADHCAI. J Dent Res 88(11):991–996. https://doi.org/10.1177/0022034509349454
Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, Park JT, Seo BM, Hu JC, Simmer JP (2008) FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am J Hum Genet 82(2):489–494. https://doi.org/10.1016/j.ajhg.2007.09.020
Wang S-K, Hu Y, Yang J, Smith CE, Richardson AS, Yamakoshi Y, Lee Y-L, Seymen F, Koruyucu M, Gencay K et al (2016) Fam83h null mice support a neomorphic mechanism for human ADHCAI. Mol Genet Genomic Med 4(1):46–67. https://doi.org/10.1002/mgg3.178
Wang SK, Hu Y, Smith CE, Yang J, Zeng C, Kim JW, Hu JC, Simmer JP (2019) The enamel phenotype in homozygous Fam83h truncation mice. Mol Genet Genomic Med 7(6):e724. https://doi.org/10.1002/mgg3.724
Nasseri S, Nikkho B, Parsa S, Ebadifar A, Soleimani F, Rahimi K, Vahabzadeh Z, Khadem-Erfan MB, Rostamzadeh J, Baban B et al (2019) Generation of Fam83h knockout mice by CRISPR/Cas9-mediated gene engineering. J Cell Biochem 120(7):11033–11043. https://doi.org/10.1002/jcb.28381
Xin W, Wang W, Man Q, Zhao Y (2017) Novel FAM83H mutations in patients with amelogenesis imperfecta. Sci Rep 7:6075. https://doi.org/10.1038/s41598-017-05208-0
Du Q, Cao L, Liu Y, Pang C, Wu S, Zheng L, Jiang W, Na X, Yu J, Wang S et al (2021) Phenotype and molecular characterizations of a family with dentinogenesis imperfecta shields type II with a novel DSPP mutation. Ann Transl Med 9(22):1672. https://doi.org/10.21037/atm-21-5369
Kawano S, Morotomi T, Toyono T, Nakamura N, Uchida T, Ohishi M, Toyoshima K, Harada H (2002) Establishment of dental epithelial cell line (HAT-7) and the cell differentiation dependent on Notch signaling pathway. Connect Tissue Res 43(2–3):409–412. https://doi.org/10.1080/03008200290000637
Bäckman B, Holm AK (1986) Amelogenesis imperfecta: prevalence and incidence in a northern Swedish county. Community Dent Oral Epidemiol 14(1):43–47. https://doi.org/10.1111/j.1600-0528.1986.tb01493.x
Yu S, Quan J, Wang X, Sun X, Zhang X, Liu Y, Zhang C, Zheng S (2018) A novel FAM83H mutation in one Chinese family with autosomal-dominant hypocalcification amelogenesis imperfecta. Mutagenesis 33(4):333–340. https://doi.org/10.1093/mutage/gey019
Simmer JP, Richardson AS, Hu YY, Smith CE, Ching-Chun HuJ (2012) A post-classical theory of enamel biomineralization and why we need one. Int J Oral Sci 4(3):129–134. https://doi.org/10.1038/ijos.2012.59
Moradian-Oldak J (2012) Protein-mediated enamel mineralization Front Biosci (Landmark Ed) 17(6):1996–2023. https://doi.org/10.2741/4034
Kuga T, Kume H, Adachi J, Kawasaki N, Shimizu M, Hoshino I, Matsubara H, Saito Y, Nakayama Y, Tomonaga T (2016) Casein kinase 1 is recruited to nuclear speckles by FAM83H and SON. Sci Rep 6:34472. https://doi.org/10.1038/srep34472
Blackburn JB, D’Souza Z, Lupashin VV (2019) Maintaining order: COG complex controls Golgi trafficking, processing, and sorting. FEBS Lett 593(17):2466–2487. https://doi.org/10.1002/1873-3468.13570
Hu CC, Hart TC, Dupont BR, Chen JJ, Sun X, Qian Q, Zhang CH, Jiang H, Mattern VL, Wright JT et al (2000) Cloning human enamelin cDNA, chromosomal localization, and analysis of expression during tooth development. J Dent Res 79(4):912–919. https://doi.org/10.1177/00220345000790040501
Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y (2004) Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol 167(5):973–983. https://doi.org/10.1083/jcb.200409077
Smith CEL, Kirkham J, Day PF, Soldani F, McDerra EJ, Poulter JA, Inglehearn CF, Mighell AJ, Brookes SJ (2017) A fourth KLK4 mutation is associated with enamel hypomineralisation and structural abnormalities. Front Physiol 29(8):333. https://doi.org/10.3389/fphys.2017.00333
Funding
We thank the patients and their family members for consenting to this research. This study was funded by grants from the National Natural Science Foundation of China (82170947) and the Applied Foundation in Science and Technology Office of Sichuan Province (2021YFG0230).
Author information
Authors and Affiliations
Contributions
JZ and QD contributed to the study conception and design. Bioinformatics and statistical analyses were performed by LC and JY. Clinical data collection and analysis were performed by QM, XH, YY and QY. Material preparation, experiment, data collection and analyses performed by YX and MM. The first draft of the manuscript was written by YX and MM, and all authors commented on previous versions of the manuscript. All authors read and approved the final.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Ethics Committee of the Sichuan Academy of Medical Sciences-Sichuan Provincial People’s Hospital (Chengdu, China) [2020 (371)]. The animal experimental protocol was approved by the Ethics Committee of West China College of Stomatology, Sichuan University, Chengdu, China (WCHSIRB-D-2021–077).
Consent to participate
Informed consent was obtained from the legal guardians of children involved in the study.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The two correspondence authors contribute equally to this work.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xie, Y., Meng, M., Cao, L. et al. Amelogenesis imperfecta in a Chinese family resulting from a FAM83H variation and the effect of FAM83H on the secretion of enamel matrix proteins. Clin Oral Invest 27, 1289–1299 (2023). https://doi.org/10.1007/s00784-022-04763-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-022-04763-9