Abstract
Objectives
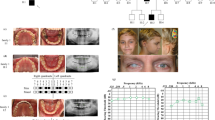
The aim of this study was the analysis of WNT10A variants in seven families of probands with various forms of tooth agenesis and self-reported family history of cancer.
Materials and methods
We enrolled 60 young subjects (aged 13 to 17) from the Czech Republic with various forms of tooth agenesis. Dental phenotypes were assessed using Planmeca ProMax 3D (Planmeca Oy, Finland) with Planmeca Romexis software (version 2.9.2) together with oral examinations. After screening PAX9, MSX1, EDA, EDAR, AXIN2 and WNT10A genes on the Illumina MiSeq platform (Illumina, USA), we further analyzed the evolutionarily highly conserved WNT10A gene by capillary sequencing in the seven families.
Results
All the detected variants were heterozygous or compound heterozygous with various levels of phenotypic expression. The most severe phenotype (oligodontia) was found in a proband who was compound heterozygous for the previously identified WNT10A variant p.Phe228Ile and a newly discovered c.748G > A variant (p.Gly250Arg) of WNT10A. The newly identified variant causes substitution of hydrophobic glycine for hydrophilic arginine.
Conclusions
We suggest that the amino acid changes in otherwise highly conserved sequences significantly affect the dental phenotype. No relationship between the presence of WNT10A variants and a risk of cancer has been found.
Clinical relevance
Screening of PAX9, MSX1, EDA, EDAR, AXIN2 and WNT10A genes in hope to elucidate the pattern of inheritance in families.


Similar content being viewed by others
References
Thesleff I (2006) The genetic basis of tooth development and dental defects. Am J Med Genet A 140:2530–2535. https://doi.org/10.1002/ajmg.a.31360
Tucker A, Sharpe PT (1999) Molecular genetics of tooth morphogenesis and patterning: the right shape in the right place. J Dent Res 78:826–834. https://doi.org/10.1177/00220345990780040201
Tucker A, Sharpe P (2004) The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet 5:499–508. https://doi.org/10.1038/nrg1380
Vieira AR, Meira R, Modesto A, Murray JC (2004) MSX1, PAX9, and TGFA contribute to tooth agenesis in humans. J Dent Res 83:723–727. https://doi.org/10.1177/154405910408300913
van der Schalk-Weide Y, Beemer FA (1994) Faber Ja, Bosman F Symptomatology of patients with oligodontia. J Oral Rehabil 21:247–261. https://doi.org/10.1111/j.1365-2842.1994.tb01141.x
Khalaf K, Miskelly J, Voge E, Macfarlane TV (2014) Prevalence of hypodontia and associated factors: a systematic review and meta-analysis. J Orthod 41:299–316. https://doi.org/10.1179/1465313314Y.0000000116
Nieminen P (2009) Genetic basis of tooth agenesis. J Exp Zool B Mol Dev Evol 312B:320–342. https://doi.org/10.1002/jez.b.21277
Polder BJ, Van’t Hof MA, Van Der Linden FPGA, Kuijpers-Jagtman AM (2004) A meta-analysis of the prevalence of dental agenesis of permanent teeth Community. Dent Oral Epidemiol 32:217–226. https://doi.org/10.1111/j.1600-0528.2004.00158.x
Jussila M, Thesleff I (2012) Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb Perspect Biol 4:a008425. https://doi.org/10.1101/cshperspect.a008425
Bailleul-Forestier I, Molla M, Verloes A, Berdal A (2008) The genetic basis of inherited anomalies of the teeth. Part 1: clinical and molecular aspects of non-syndromic dental disorders. Eur J Med Genet 51:273–291. https://doi.org/10.1016/j.ejmg.2008.02.009
Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE (1996) A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet 13:417–421. https://doi.org/10.1038/ng0896-417
Šerý O, Bonczek O, Hloušková A, Černochová P, Vaněk J, Míšek I, Krejčí P, Izakovičová Hollá L (2015) A screen of a large Czech cohort of oligodontia patients implicates a novel mutation in the PAX9 gene. Eur J Oral Sci 123:65–71. https://doi.org/10.1111/eos.12170
Bonczek O, Balcar VJ, Šerý O (2017) PAX9 gene mutations and tooth agenesis: A review. Clin Genet 92:467–476. https://doi.org/10.1111/cge.12986
Song S, Han D, Qu H, Gong Y, Wu H, Zhang X, Zhong N, Feng H (2009) EDA gene mutations underlie non-syndromic oligodontia. J Dent Res 88:126–131. https://doi.org/10.1177/0022034508328627
Bergendal B, Klar J, Stecksén-Blicks C, Norderyd J, Dahl N (2011) Isolated oligodontia associated with mutations in EDARADD, AXIN2, MSX1, and PAX9 genes. Am J Med Genet A 155A:1616–1622. https://doi.org/10.1002/ajmg.a.34045
Mostowska A, Biedziak B, Zadurska M, Matuszewska-Trojan S, Jagodziński PP (2015) WNT10A coding variants and maxillary lateral incisor agenesis with associated dental anomalies. Eur J Oral Sci 123:1–8. https://doi.org/10.1111/eos.12165
Peifer M, Polakis P (2000) Wnt signaling in oncogenesis and embryogenesis - a look outside the nucleus. Science 287:1606–1609. https://doi.org/10.1126/science.287.5458.1606
Yuan Q, Zhao M, Tandon B, Maili L, Liu X, Zhang A, Baugh EH, Tran T, Rm S, Hecht JT, Swindell EC, Wagner DS, Letra A (2017) Role of WNT10A in failure of tooth development in humans and zebrafish. Mol Genet Genomic Med 5:730–741. https://doi.org/10.1002/mgg3.332
Miller JR (2002) The Wnts. Genome Biol 3(1):reviews3001.1–reviews3001.15. https://doi.org/10.1186/gb-2001-3-1-reviews3001
Bodine PVN, Komm BS (2006) Wnt signaling and osteoblastogenesis. Rev Endocr Metab Disord 7:33–39. https://doi.org/10.1007/s11154-006-9002-4
Van Amerongen R, Nusse R (2009) Towards an integrated view of Wnt signaling in development. Development 136:3205–3214. https://doi.org/10.1242/dev.033910
Moon RT, Shah K (2002) Developmental biology: signalling polarity. Nature 417:239–240. https://doi.org/10.1038/417239a
Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier W, Paus R, Piccolo S, Mikkola ML, Morrisey EE, Overbeek PA, Scheidereit C, Millar SE, Schmidt-Ullrich R (2009) Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Dev Cell 17:49–61. https://doi.org/10.1016/j.devcel.2009.05.011
Mostowska A, Hozyasz KK, Biedziak B, Wojcicki P, Lianeri M, Jagodzinski PP (2012) Genotype and haplotype analysis of WNT genes in non-syndromic cleft lip with or without cleft palate. Eur J Oral Sci 120:1–8. https://doi.org/10.1111/j.1600-0722.2011.00938.x
Mues G, Bonds J, Xiang L, Vieira AR, Seymen F, Klein O, D’souza RN (2014) The WNT10A gene in ectodermal dysplasias and selective tooth agenesis. Am J Med Genet A 164A:2455–2460. https://doi.org/10.1002/ajmg.a.36520
Liu F, Millar SE (2010) Wnt/beta-catenin signaling in oral tissue development and disease. J Dent Res 89:318–330. https://doi.org/10.1177/0022034510363373
Tamura M, Nemoto E, Sato MM, Nakashima A, Shimauchi H (2010) Role of the Wnt signaling pathway in bone and tooth. Front Biosci (Elite Ed) 2:1405–1413. https://doi.org/10.2741/e201
Cadigan KM, Nusse R (1997) Wnt signaling: a common theme in animal development. Genes Dev 11:3286–3305. https://doi.org/10.1101/gad.11.24.3286
Wodarz A, Nusse R (1998) Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol 14:59–88. https://doi.org/10.1146/annurev.cellbio.14.1.59
Bohring A, Stamm T, Spaich C, Haase C, Spree K, Hehr U, Hoffmann M, Ledig S, Sel S, Wieacker P, Röpke A (2009) WNT10A mutations are a frequent cause of a broad spectrum of ectodermal dysplasias with sex-biased manifestation pattern in heterozygotes. Am J Hum Genet 85:97–105. https://doi.org/10.1016/j.ajhg.2009.06.001
Wang J, Shackleford GM (1996) Murine Wnt10a and Wnt10b: cloning and expression in developing limbs, face and skin of embryos and in adults. Oncogene 13:1537–1544
Dassule HR, Mcmahon AP (1998) Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev Biol 202:215–227. https://doi.org/10.1006/dbio.1998.8992
Millar SE, Willert K, Salinas PC, Roelink H, Nusse R, Sussman DJ, Barsh GS (1999) WNT signaling in the control of hair growth and structure. Dev Biol 207:133–149. https://doi.org/10.1006/dbio.1998.9140
Andl T, Reddy ST, Gaddapara T, Millar SE (2002) WNT signals are required for the initiation of hair follicle development. Dev Cell 2:643–653. https://doi.org/10.1016/s1534-5807(02)00167-3
Bailleul-Forestier I, Berdal A, Vinckier F, De Ravel T, Fryns JP, Verloes A (2008) The genetic basis of inherited anomalies of the teeth. Part 2: syndromes with significant dental involvement. Eur J Med Genet 51:383–408. https://doi.org/10.1016/j.ejmg.2008.05.003
Cluzeau C, Hadj-Rabia S, Jambou M, Mansour S, Guigue P, Masmoudi S, Bal E, Chassaing N, Vincent MC, Viot G, Clauss F, Manière MC, Toupenay S, Le Merrer M, Lyonnet S, Cormier-Daire V, Amiel J, Faivre L, de Prost Y, Munnich A, Bonnefont JP, Bodemer C, Smahi A (2011) Only four genes (EDA1, EDAR, EDARADD, and WNT10A) account for 90% of hypohidrotic/anhidrotic ectodermal dysplasia cases. Hum Mutat 32:70–72. https://doi.org/10.1002/humu.21384
Wedgeworth EK, Nagy N, White JML, Pembroke AC, Mcgrath JA (2011) Intra-familial variability of ectodermal defects associated with WNT10A mutations. Acta Derm Venereol 91:346–347. https://doi.org/10.2340/00015555-1028
Van Den Boogaard MJ, Créton M, Bronkhorst Y, Van Der Hout A, Hennekam E, Lindhout D, Cune M, Van Amstel HKP (2012) Mutations in WNT10A are present in more than half of isolated hypodontia cases. J Med Genet 49:327–331. https://doi.org/10.1136/jmedgenet-2012-100750
Mostowska A, Biedziak B, Zadurska M, Dunin-Wilczynska I, Lianeri M, Jagodzinski PP (2013) Nucleotide variants of genes encoding components of the Wnt signalling pathway and the risk of non-syndromic tooth agenesis. Clin Genet 84:429–440. https://doi.org/10.1111/cge.12061
Plaisancié J, Bailleul-Forestier I, Gaston V, Vaysse F, Lacombe D, Holder-Espinasse M, Abramowicz M, Coubes C, Plessis G, Faivre L, Demeer B, Vincent-Delorme C, Dollfus H, Sigaudy S, Guillén-Navarro E, Verloes A, Jonveaux P, Martin-Coignard D, Colin E, Bieth E, Calvas P, Chassaing N (2013) Mutations in WNT10A are frequently involved in oligodontia associated with minor signs of ectodermal dysplasia. Am J Med Genet A 161A:671–678. https://doi.org/10.1002/ajmg.a.35747
Song S, Zhao R, He H, Zhang J, Feng H, Lin L (2014) WNT10A variants are associated with non-syndromic tooth agenesis in the general population. Hum Genet 133:117–124. https://doi.org/10.1007/s00439-013-1360-x
Arzoo PS, Klar J, Bergendal B, Norderyd J, Dahl N (2014) WNT10A mutations account for ¼ of population-based isolated oligodontia and show phenotypic correlations. Am J Med Genet A 164A:353–359. https://doi.org/10.1002/ajmg.a.36243
Clevers H (2006) Wnt/beta-catenin signaling in development and disease. Cell 127:469–480. https://doi.org/10.1016/j.cell.2006.10.018
Li J, Zhang Z, Wang L, Zhang Y (2019) The oncogenic role of Wnt10a in colorectal cancer through activation of canonical Wnt/β-catenin signaling. Oncol Lett 17:3657–3664. https://doi.org/10.3892/ol.2019.10035
Bonczek O, Krejci P, Izakovicova-Holla L, Cernochova P, Kiss I, Vojtesek B (2021) Tooth agenesis: What do we know and is there a connection to cancer? Clin Genet 99:493–502. https://doi.org/10.1111/cge.13892
Jia S, Zhou J, Fanelli C, Wee Y, Bonds J, Schneider P, Mues G, D’Souza RN (2017) Small-molecule Wnt agonists correct cleft palates in Pax9 mutant mice in utero. Development 144:3819–3828. https://doi.org/10.1242/dev.157750
Yu M, Wong SW, Han D, Cai T (2019) Genetic analysis: Wnt and other pathways in nonsyndromic tooth agenesis. Oral Dis 25:646–651. https://doi.org/10.1111/odi.12931
Roche: Sequencing Solutions Technical Note: How To Evaluate NimbleGen SeqCap EZ Target Enrichment Data Roche Diagnostics: Mannheim. http://netdocs.roche.com/DDM/Effective/07187009001_RNG_SeqCap-EZ_TchNote_Eval-data_v2.1.pdf Accessed 2 October 2020
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–17660. https://doi.org/10.1093/bioinformatics/btp324
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. https://doi.org/10.1093/bioinformatics/btr509
R. CoreTeam (2015) R:A language and environment for statistical computing. Available: https://www.R-project.org/. Accessed 28 September 2020
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. https://doi.org/10.1038/nbt.1754
Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. https://doi.org/10.1093/bib/bbs017
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. https://doi.org/10.1038/nmeth0410-248
Arte S, Parmanen S, Pirinen S, Alaluusua S, Nieminen P (2013) Candidate gene analysis of tooth agenesis identifies novel mutations in six genes and suggests significant role for WNT and EDA signaling and allele combinations. PLoS One 8:e73705. https://doi.org/10.1371/journal.pone.0073705
Kantaputra P, Sripathomsawat W (2011) WNT10A and isolated hypodontia. Am J Med Genet A 155A:1119–1122. https://doi.org/10.1002/ajmg.a.33840
Zhang J, Tian XJ, Xing J (2016) Signal Transduction Pathways of EMT Induced by TGF-β, SHH, and WNT and Their Crosstalks. J Clin Med 5:41. https://doi.org/10.3390/jcm5040041
Heise RL, Stober V, Cheluvaraju C, Hollingsworth JW, Garantziotis S (2011) Mechanical stretch induces epithelial-mesenchymal transition in alveolar epithelia via hyaluronan activation of innate immunity. J Biol Chem 286:17435–17444. https://doi.org/10.1074/jbc.M110.137273
Hlouskova A, Bielik P, Bonczek O, Balcar VJ, Šerý O (2017) Mutations in AXIN2 gene as a risk factor for tooth agenesis and cancer: A review. Neuro Endocrinol Lett 38:131–137
He H, Han D, Feng H, Qu H, Song S, Bai B, Zhang Z (2013) Involvement of and interaction between WNT10A and EDA mutations in tooth agenesis cases in the Chinese population. PLoS One 8:e80393. https://doi.org/10.1371/journal.pone.0080393
Suomalainen M, Thesleff I (2010) Patterns of Wnt pathway activity in the mouse incisor indicate absence of Wnt/beta-catenin signaling in the epithelial stem cells. Dev Dyn 239:364–372. https://doi.org/10.1002/dvdy.22106
Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC (2012) Structural basis of Wnt recognition by Frizzled. Science 337:59–64. https://doi.org/10.1126/science.1222879
Nawaz S, Klar J, Wajid M, Aslam M, Tariq M, Schuster J, Baig SM, Dahl N (2009) WNT10A missense mutation associated with a complete odonto-onycho-dermal dysplasia syndrome. Eur J Hum Genet 17:1600–1605. https://doi.org/10.1038/ejhg.2009.81
Tardieu C, Jung S, Niederreither K, Prasad M, Hadj-Rabia S, Philip N, Mallet A, Consolino E, Sfeir E, Noueiri B, Chassaing N, Dollfus H, Manière MC, Bloch-Zupan A, Clauss F (2017) Dental and extra-oral clinical features in 41 patients with WNT10A gene mutations: A multicentric genotype-phenotype study. Clin Genet 92:477–486. https://doi.org/10.1111/cge.12972
Acknowledgements
We would like to thank all the subjects who participated in the research. The authors thank Dr. Philip J. Coates (RECAMO, MMCI, Brno, Czech Republic) for English language editing.
Funding
This work was funded by grants of the IGA MH CZ n.: NT/11420–6/2010, the European Regional Development Fund-Project ENOCH (No. CZ.02.1.01/0.0/0.0/16_019/0000868), MH CZ-DRO (MMCI, 00209805) and AZV CR NU20-06–00189.
Author information
Authors and Affiliations
Contributions
All authors made substantial contribution to the conception and design of the manuscript. P.B. and O.B. drafted manuscript, T.Z. analyzed data from NGS sequencing, P.B. and J.L. performed NGS sequencing, P.K., J.S. and J.V. performed sampling and clinical investigations, J.V., L.I.H. and O.S. conceptualized research and obtained fundings, B.V. and V.J.B. edited and supervised manuscript, and O.S. supervised laboratory analyses and final manuscript. All authors agree to be accountable for all aspects of the study design and its content. All authors approved the final submitted version.
Corresponding author
Ethics declarations
Ethical approval
The study protocol and informed consents were reviewed and approved by the ethical committee of Faculty of Medicine and Dentistry, Olomouc and Faculty of Medicine, Brno. The study was performed in accordance with the Declaration of Helsinki. Care was taken to follow the letter and spirit of the Declaration of Helsinki—ethical principles for medical research involving human subjects.
Informed consent
Written informed consent (No: Fm-L009-001-ZUBNI-014) was obtained from all participating human subjects.
Conflict of interest
All the authors declare that they have no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bielik, P., Bonczek, O., Krejčí, P. et al. WNT10A variants: following the pattern of inheritance in tooth agenesis and self-reported family history of cancer. Clin Oral Invest 26, 7045–7055 (2022). https://doi.org/10.1007/s00784-022-04664-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-022-04664-x