Abstract
Objectives
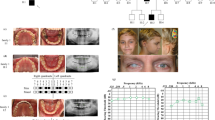
KDF1 is a recently identified gene related to tooth development, but it has been little studied. To date, only three cases have been reported in which KDF1 mutations are related to tooth development, including two ectodermal dysplasia cases accompanied by tooth loss and one non-syndromic case with tooth agenesis. However, no KDF1 mutations have been reported as associated with non-syndromic anodontia. Here, the aim was to investigate the genetic etiology of this condition and explore the functional role of a novel KDF1 mutation in a Chinese patient with non-syndromic anodontia.
Materials and methods
Pathogenic variants were identified by whole-exome and Sanger sequencing. Meanwhile, we conducted a literature review of the reported KDF1 mutations and performed an in vitro functional analysis of four anodontia-causing KDF1 mutations (one novel and three known).
Results
We identified a novel de novo missense mutation (c.911 T > A, p.I304N) in the KDF1 gene in a Chinese patient with severe non-syndromic anodontia. In vitro functional studies showed altered mRNA and protein expression levels of the mutant KDF1.
Conclusions
Our results are the first report of KDF1 missense mutation causing non-syndromic anodontia.
Clinical relevance
This study not only further supports the important role of KDF1 in non-syndromic congenital anodontia, but also expands the spectrum of KDF1 mutations and will contribute to the genetic diagnosis and counselling of families with anodontia.





Similar content being viewed by others
References
de Smalen A, van Nunen DPF, Hermus RR, Ongkosuwito EM, van Wijk AJ, Griot JPWD, Breugem CC, Kramer GJC (2017) Permanent tooth agenesis in non-syndromic Robin sequence and cleft palate: prevalence and patterns. Clin Oral Invest 21(7):2273–2281. https://doi.org/10.1007/s00784-016-2020-z
Salvi A, Giacopuzzi E, Bardellini E, Amadori F, Ferrari L, De Petro G, Borsani G, Majorana A (2016) Mutation analysis by direct and whole exome sequencing in familial and sporadic tooth agenesis. Int J Mol Med 38(5):1338–1348. https://doi.org/10.3892/ijmm.2016.2742
Acikgoz A, Kademoglu O, Elekdag-Turk S, Karagoz F (2007) Hypohidrotic ectodermal dysplasia with true anodontia of the primary dentition. Quintessence Int 38(10):853–858
Salas-Alanis JC, Cepeda-Valdes R, Gonzalez-Santos A, Amaya-Guerra M, Kurban M, Christiano AM (2011) Mutation in the ED1, Ala349Thr in a patient with X-linked hypohidrotic ectodermal dysplasia. Rev Med Chile 139(12):1601–1604
Fauzi NH, Ardini YD, Zainuddin Z, Lestari W (2018) A review on non-syndromic tooth agenesis associated with PAX9 mutations. Jpn Dent Sci Rev 54(1):30–36. https://doi.org/10.1016/j.jdsr.2017.08.001
Wang J, Sun K, Shen Y, Xu YZ, Xie J, Huang RH, Zhang YM, Xu CY, Zhang X, Wang RR (2016) DNA methylation is critical for tooth agenesis: implications for sporadic non-syndromic anodontia and hypodontia. Sci Rep-Uk 6:19162. https://doi.org/10.1038/srep19162
Zhou YJ, Liao YD, Zhang CY, Liu JX, Wang W, Huang JM, Du QQ, Liu TY, Zou QJ, Huang H (2021) TAB2 promotes the stemness and biological functions of cervical squamous cell carcinoma cells. Stem Cells Int 2021:6550388. https://doi.org/10.1155/2021/6550388
Zhang LT, Yu M, Sun K, Fan ZZ, Liu HC, Feng HL, Liu Y, Han D (2021) Rare phenotype: Hand preaxial polydactyly associated with LRP6-related tooth agenesis in humans. Npj Genom Med 6(1):93. https://doi.org/10.1038/s41525-021-00262-0
Doolan BJ, Onoufriadis A, Kantaputra P, McGrath JA (2021) WNT10A, dermatology and dentistry. Brit J Dermatol 185(6):1105–1111. https://doi.org/10.1111/bjd.20601
Han Y, Wang XL, Zheng LY, Zhu TT, Li YW, Hong JQ, Xu CC, Wang PG, Gao M (2020) Pathogenic EDA mutations in Chinese Han families with hypohidrotic ectodermal dysplasia and genotype-phenotype: a correlation analysis. Front Genet 4(11):21. https://doi.org/10.3389/fgene.2020.00021
Okita T, Asano N, Yasuno S, Shimomura Y (2019) Functional studies for a dominant mutation in the EDAR gene responsible for hypohidrotic ectodermal dysplasia. J Dermatol 46(8):710–715. https://doi.org/10.1111/1346-8138.14983
Chassaing N, Cluzeau C, Bal E, Guigue P, Vincent MC, Viot G, Ginisty D, Munnich A, Smahi A, Calvas P (2010) Mutations in EDARADD account for a small proportion of hypohidrotic ectodermal dysplasia cases. Brit J Dermatol 162(5):1044–1048. https://doi.org/10.1111/j.1365-2133.2010.09670.x
Andreoni F, Sgattoni C, Bencardino D, Simonetti O, Forabosco A, Magnani M (2020) Missense mutations in EDA and EDAR genes cause dominant syndromic tooth agenesis. Mol Genet Genom Med 9(1):e1555. https://doi.org/10.1002/mgg3.1555
van den Boogaard MJ, Creton M, Bronkhorst Y, van der Hout A, Hennekam E, Lindhout D, Cune M, van Amstel HKP (2012) Mutations in WNT10A are present in more than half of isolated hypodontia cases. J Med Genet 49(5):327–331. https://doi.org/10.1136/jmedgenet-2012-100750
Shamseldin HE, Khalifa O, Binamer YM, Almutawa A, Arold ST, Zaidan H, Alkuraya FS (2017) KDF1, encoding keratinocyte differentiation factor 1, is mutated in a multigenerational family with ectodermal dysplasia. Hum Genet 136(1):99–105. https://doi.org/10.1007/s00439-016-1741-z
Manaspon C, Thaweesapphithak S, Osathanon T, Suphapeetiporn K, Porntaveetus T, Shotelersuk V (2019) A novel de novo mutation substantiates KDF1 as a gene causing ectodermal dysplasia. Brit J Dermatol 181(2):419–420. https://doi.org/10.1111/bjd.18007
Zeng BH, Lu H, Xiao X, Yu XL, Li SJ, Zhu L, Yu DS, Zhao W (2019) KDF1 is a novel candidate gene of non-syndromic tooth agenesis. Arch Oral Biol 97:131–136. https://doi.org/10.1016/j.archoralbio.2018.10.025
Santos EM, Paula JF, Motta PM, Heinemann MB, Leite RC, Haddad JP, Del Puerto HL, Reis JK (2010) Comparison of three methods of DNA extraction from peripheral blood mononuclear cells and lung fragments of equines. Genet Mol Res 9(3):1591–8. https://doi.org/10.4238/vol9-3gmr818
Lee S, Kong Y, Weatherbee SD (2013) Forward genetics identifies Kdf1/1810019J16Rik as an essential regulator of the proliferation-differentiation decision in epidermal progenitor cells. Dev Biol 383(2):201–213. https://doi.org/10.1016/j.ydbio.2013.09.022
Li YY, Tang LF, Yue JP, Gou XW, Lin AN, Weatherbee SD, Wu XY (2020) Regulation of epidermal differentiation through KDF1-mediated deubiquitination of IKK alpha. Embo Rep 21(5):e48566. https://doi.org/10.15252/embr.201948566.
Zheng JM, Gan MF, Yu HY, Ye LX, Yu QX, Xia YH, Zhou HX, Bao JQ, Guo YQ (2021) KDF1, a novel tumor suppressor in clear cell renal cell carcinoma. Front Oncol 11:686678. https://doi.org/10.3389/fonc.2021.686678
Funding
National Natural Science Foundation of China (31970558, 82170920), National Key S&T Special Projects (2021YFC1005301), Natural Science Foundation of Guangdong Province of China (2018B030311033), Health Department of Guangxi Zhuang Autonomous Region (Z20200678), Shenzhen Fundamental Research Program (JCYJ20180228164057158).
Author information
Authors and Affiliations
Contributions
Yuhua Pan: conceptualization; methodology; formal analysis; writing—original draft; writing—review and editing. Sheng Yi: software, investigation, resources, funding acquisition. Dong Chen and Xinya Du: methodology, formal analysis, software. Xinchen Yao: resources, data curation. Fei He: visualization, supervision, project administration, funding acquisition. Fu Xiong: validation; writing—review and editing; project administration; funding acquisition.
Corresponding authors
Ethics declarations
Ethical approval
Approval was obtained from the Ethics Committee of Nanfang Hospital, an affiliate of Southern Medical University (No. NFEC-2021–1071). The procedures used in this study adhere to the tenets of the Declaration of Helsinki.
Informed consent
Written informed consent was obtained from all participants or their guardians.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pan, Y., Yi, S., Chen, D. et al. Identification of a novel missense heterozygous mutation in the KDF1 gene for non-syndromic congenital anodontia. Clin Oral Invest 26, 5171–5179 (2022). https://doi.org/10.1007/s00784-022-04485-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-022-04485-y