Abstract
Background
Molecular biological techniques such as the polymerase chain reaction (PCR) and DNA microarray are used for the detection/identification of microorganisms; however, few reports have discussed the clinical utility of microarray analysis for identification of causative organisms of osteoarticular infections. It is important to examine the utility of PCR amplification followed by analysis of DNA microarray carrying specific oligonucleotides.
Methods
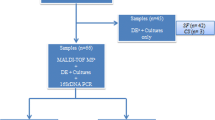
This study included 101 biological samples obtained from 96 patients who underwent conservative and/or surgical treatment for osteoarticular infections. In this double-blind comparative study, routine conventional testing and the research groups were unaware of each other’s interpretation until identical specimens were identified by culture and microarray analysis.
Results
Results of PCR microarray analysis were positive for 25 samples and negative for the remaining 76 samples within 24 h, and the results of the cultures (available after a mean of 3.54 days) were positive in 26 samples and negative for the remaining 75 samples. The sensitivity of microarray analysis was 84.6% (22/26) and specificity was 88.0% (22/25). Discrepant results were identified in seven samples, including a negative culture and a positive microarray in three cases and a positive culture and a negative microarray in four other cases.
Conclusions
The PCR microarray analysis is complementary to routine cultures in identifying causative microorganisms and should be used in patients with highly suspected infections and negative bacterial culture and in patients who require prompt diagnosis and early initiation of antibiotic therapy.
Similar content being viewed by others
References
Lew DP, Waldvogel FA. Osteomyelitis. Lancet 2004;364:369–379.
Shirtliff ME, Mader JT. Acute septic arthritis. Clin Microbiol Rev 2002;15:527–544.
Zimmerli W, Trampuz A, Ochsner PE. Prosthetic-joint infections. N Engl J Med 2004;351:1645–1654.
Gupta MN, Sturrock RD, Field M. Prospective comparative study of patients with culture proven and high suspicion of adult onset septic arthritis. Ann Rheum Dis 2003;62:327–331.
Kaandorp CJ, Krijnen P, Moens HJ, Habbema JD, van Schaardenburg D. The outcome of bacterial arthritis: a prospective community-based study. Arthritis Rheum 1997;40:884–892.
Uchida K, Kobayashi S, Sato R, Nakajima H, Yayama T, Kokubo Y, et al. Anterior spinal artery syndrome complicating massive paravertebral abscess. J Orthop Sci 2008;13:85–88.
Tunney MM, Patrick S, Curran MD, Ramage G, Hanna D, Nixon JR, et al. Detection of prosthetic hip infection at revision arthropathy by immunofluorescence microscopy and PCR amplification of the bacterial 16S rRNA gene. J Clin Microbiol 1999;37:3281–3290.
Patel JB. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol Diagn 2001;6:313–321.
Yang S, Rothman RE. PCR-based diagnostics for infectious disease: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis 2004;4:337–348.
Fenollar F, Roux V, Stein A, Drancourt M, Raoult D. Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J Clin Microbiol 2006;44:1018–1028.
Kobayashi N, Bauer TW, Togawa D, Lieberman IH, Sakai H, Fujishiro T, et al. A molecular gram stain using broad range PCR and pyrosequencing technology: a potentially useful tool for diagnosing orthopaedic infections. Diagn Mol Pathol 2005;14:83–89.
Anthony RM, Brown TJ, French GL. DNA array technology and diagnostic microbiology. Expert Rev Mol Diagn 2001;1:30–38.
Tillib SV, Mirzabekov AD. Advances in the analysis of DNA sequence variations using oligonucleotide microchip technology. Curr Opin Biotechnol 2001;12:53–58.
Anthony RM, Brown TJ, French GL. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J Clin Microbiol 2000;38:781–788.
Mitterer G, Huber M, Leidinger E, Kirisits C, Lubitz W, Muellar MW, Schmidt WM. Microarray-based identification of bacteria in clinical samples by solid-phase PCR amplification of 23S ribosomal DNA sequences. J Clin Microbiol 2004;42:1048–1057.
Hall GS. Molecular applications in the clinical laboratory. In: Mahon CR, Manuselis G, editors. Diagnostic microbiology. 2nd edn. Philadelphia: Saunders; 2000. p. 191–210.
Al Zahrani K, Al Jahdali H, Poirier L, René P, Gennaro ML, Menzies D. Accuracy and utility of commercially available amplification and serologic tests for the diagnosis of minimal pulmonary tuberculosis. Am J of Respir Crit Care Med 2000;162:1323–1329.
Berndt C, Müller U, Bergmann F, Schmitt U, Kaiser R, Müller C. Comparison between a nucleic acid sequence-based amplification and branched DNA test for quantifying HIV RNA load in blood plasma. J Virol Methods 2000;89:177–181.
Uchida K, Baba H, Wada M, Maezawa Y, Kikukawa Y, Imura S. Atypical mycobacterium osteomyelitis of the humerus in a child with chronic granulomatous disease. J Neurol Orthop Med Surg 1996;17:127–129.
Jordan JA, Durso MB. Comparison of 16S rRNA gene PCR and BACTEC 9240 for detection of neonatal bacteremia. J Clin Microbiol 2000;38:2574–2578.
Kobayashi N, Procop GW, Krebs V, Kobayashi H, Bauer TW. Molecular identification of bacteria from aseptically loose implants. Clin Orthop 2008;466:1716–1725.
Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol 1991;173:697–703.
Carroll NM, Jaeger EE, Choudhury S, Dunlop AA, Matheson MM, Adamson P, et al. Detection of and discrimination between gram-positive and gram-negative bacteria in intraocular samples by using nested PCR. J Clin Microbiol 2000;38:1753–1757.
Xu J, Millar BC, Moore JE, Murphy K, Webb H, Fox AJ, et al. Employment of broad-range 16S rRNA PCR to detect aetiological agents of infection from clinical specimens in patients with acute meningitis: rapid separation of 16S rRNA PCR amplicon without the need for cloning. J Appl Microbiol 2003;94:197–206.
Tsai JC, Teng LJ, Hsueh PR. Direct detection of bacterial pathogens in brain abscesses by polymerase chain reaction amplification and sequencing of partial 16S ribosomal deoxyribonucleic acid fragments. Neurosurgery 2004;55:1154–1162.
Wilbrink B, van der Heijden IM, Schouls LM, van Embden JD, Hazes JM, Breedveld FC, et al. Detection of bacterial DNA in joint samples from patients with undifferentiated arthritis and reactive arthritis, using polymerase chain reaction with universal 16S ribosomal RNA primers. Arthritis Rheum 1998;41:535–543.
Jackson CJ, Barton RC, Evans EG. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. J Clin Microbiol 1999;37:931–936.
Morace G, Sanguinetti M, Posteraro B, Lo Cascio L, Fadda G. Identification of various medically important Candida species in clinical specimens by PCR-restriction enzyme analysis. J Clin Microbiol 1997;35:667–672.
Henry T, Iwen PC, Hinrichs SH. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J Clin Microbiol 2000;38:1510–1515.
Fujita S, Senda Y, Nakaguchi S, Hashimoto T. Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J Clin Microbiol 2001;39:3617–3622.
Turenne CY, Sanche SE, Hoban DJ, Karlowsky JA, Kabani AM. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol 1999;37:1846–1851.
Author information
Authors and Affiliations
About this article
Cite this article
Uchida, K., Yayama, T., Kokubo, Y. et al. Direct detection of pathogens in osteoarticular infections by polymerase chain reaction amplification and microarray hybridization. J Orthop Sci 14, 471–483 (2009). https://doi.org/10.1007/s00776-009-1373-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00776-009-1373-4