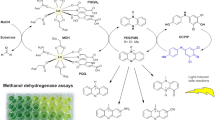
Abstract
CO dehydrogenase (CODH) is an environmentally crucial bacterial enzyme that oxidizes CO to CO2 at a Mo–Cu active site. Despite the close to atomic resolution structure (1.1 Å), significant uncertainties have remained with regard to the protonation state of the water-derived equatorial ligand coordinated at the Mo-center, as well as the nature of intermediates formed during the catalytic cycle. To address the protonation state of the equatorial ligand, we have developed a realistic in silico QM model (~179 atoms) containing structurally essential residues surrounding the active site. Using our QM model, we examined each plausible combination of redox states (MoVI–CuI, MoV–CuII, MoV–CuI, and MoIV–CuI) and Mo-coordinated equatorial ligands (O2−, OH−, H2O), as well as the effects of second-sphere residues surrounding the active site. Herein, we present a refined computational model for the Mo(VI) state in which Glu763 acts as an active site base, leading to a MoO2-like core and a protonated Glu763. Calculated structural and spectroscopic data (hyperfine couplings) are in support of a MoO2-like core in agreement with XRD data. The calculated two-electron reduction potential (E = −467 mV vs. SHE) is in reasonable agreement with the experimental value (E = −558 mV vs. SHE) for the redox couple comprising an equatorial oxo ligand and protonated Glu763 in the MoVI–CuI state and an equatorial water in the MoIV–CuI state. We also suggest a potential role of second-sphere residues (e.g., Glu763, Phe390) based on geometric changes observed upon exclusion of these residues in the most plausible oxidized states.





Similar content being viewed by others
References
Mörsdorf G, Frunzke K, Gadkari D, Meyer O (1992) Biodegradation 3:61–82
Moxley JM, Smith KA (1998) Soil Biol Biochem 30:65–79
Wilcoxen J, Hille R (2013) J Biol Chem 288:36052–36060
Dobbek H, Gremer L, Kiefersauer R, Huber R, Meyer O (2002) Proc Natl Acad Sci 99:15971–15976
Gnida M, Ferner R, Gremer L, Meyer O, Meyer-Klaucke W (2003) Biochemistry 42:222–230
Zhang B, Hemann CF, Hille R (2010) J Biol Chem 285:12571–12578
Shanmugam M, Wilcoxen J, Habel-Rodriguez D, Cutsail GE 3rd, Kirk ML, Hoffman BM, Hille R (2013) J Am Chem Soc 135:17775–17782
Siegbahn PE, Shestakov AF (2005) J Comput Chem 26:888–898
Stein BW, Kirk ML (2015) J Biol Inorg Chem 20:183–194
Neese F (2012) Wiley Interdiscip Rev Comput Mol Sci 2:73–78
Klamt A, Schuurmann G (1993) J Chem Soc Perkin Trans 2:799–805
Becke AD (1988) Phys Rev A 38:3098–3100
Perdew JP (1986) Phys Rev B 33:8822–8824
Bühl M, Kabrede H (2006) J Chem Theory Comput 2:1282–1290
Pantazis DA, Chen X-Y, Landis CR, Neese F (2008) J Chem Theory Comput 4:908–919
Neese F (2003) J Comput Chem 24:1740–1747
Weigend F (2006) Phys Chem Chem Phys 8:1057–1065
Bykov D, Petrenko T, Izsák R, Kossmann S, Becker U, Valeev E, Neese F (2015) Mol Phys 113:1961–1977
Knizia G (2013) J Chem Theory Comput 9:4834–4843
Becke AD (1993) J Chem Phys 98:5648–5652
Staroverov VN, Scuseria GE, Tao J, Perdew JP (2003) J Chem Phys 119:12129–12137
Izsak R, Neese F (2011) J Chem Phys 135:144105
Neese F, Wennmohs F, Hansen A, Becker U (2009) Chem Phys 356:98–109
Palascak MW, Shields GC (2004) J Phys Chem A 108:3692–3694
Sinnecker S, Neese F (2006) J Comput Chem 27:1463–1475
Rokhsana D, Dooley DM, Szilagyi RK (2008) J Biol Inorg Chem 13:371–383
Bjornsson R, Lima FA, Spatzal T, Weyhermuller T, Glatzel P, Bill E, Einsle O, Neese F, DeBeer S (2014) Chem Sci 5:3096–3103
Gourlay C, Nielsen DJ, White JM, Knottenbelt SZ, Kirk ML, Young CG (2006) J Am Chem Soc 128:2164–2165
Hille R, Dingwall S, Wilcoxen J (2015) J Biol Inorg Chem 20:243–251
Acknowledgments
This research was funded by generous financial contributions from Whitman College and the M. J. Murdock Charitable Trust. Special thanks to Dr. Robert Szilagyi (Montana State University, Bozeman, MT) for his tremendous assistance in setting up the computational server at Whitman College, and for providing comments and feedback during the preparation of this manuscript. We gratefully acknowledge the Max Planck Society for financial support of this work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rokhsana, D., Large, T.A.G., Dienst, M.C. et al. A realistic in silico model for structure/function studies of molybdenum–copper CO dehydrogenase. J Biol Inorg Chem 21, 491–499 (2016). https://doi.org/10.1007/s00775-016-1359-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-016-1359-6