Abstract
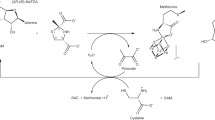
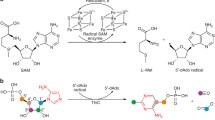
4-Hydroxyphenylacetate decarboxylase-activating enzyme (4Hpad-AE) uses S-adenosylmethionine (SAM or AdoMet) and a [4Fe-4S]2+/+ cluster (RS cluster) to generate a stable glycyl radical on the decarboxylase. 4Hpad-AE might bind up to two auxiliary [4Fe-4S] clusters coordinated by a ferredoxin-like insert C-terminal to the RS cluster-binding motif. Except for the AEs of pyruvate formate-lyase and anaerobic ribonucleotide reductase, all glycyl radical-activating enzymes possess a similar ferredoxin-like domain, whose functional role is still poorly understood. To assess the role of the putative ferredoxin clusters from 4Hpad-AE, we combined biochemical and spectroscopic methods to characterize a truncated version of the protein (Δ66-AE) devoid of the ferredoxin-like domain. We found that Δ66-AE is stable, harbors a fully active RS cluster and can activate the decarboxylase. From the similar cleavage rates for S-adenosylmethionine of Δ66-AE and wild-type AE, we infer the reactivity of the RS cluster is unperturbed by the absence of the ferredoxin-like domain. Thus, the auxiliary clusters are not required as electron conduit to the RS cluster for effective reductive cleavage of SAM. The activation of the decarboxylase by Δ66-AE is almost as fast as with wild-type AE, but the generated glycyl radical is short living. We postulate that the ferredoxin-like domain is not required for SAM-dependent glycyl radical generation in the decarboxylase, but is necessary for producing a lasting glycyl radical.






Similar content being viewed by others
Abbreviations
- Ferredoxin clusters:
-
Auxiliary [4Fe-4S] clusters coordinated by the ferredoxin-like domain
- Nrd-AE:
-
Class-III ribonucleotide reductase-activating enzyme
- EPR:
-
Electron paramagnetic resonance
- GRE:
-
Glycyl radical enzyme
- HPLC:
-
High-performance liquid chromatography
- Δ66-AE:
-
4Hpad-AE variant devoid of the 2x[4Fe-4S] ferredoxin-like domain
- 4Hpad-AE:
-
4-Hydroxyphenylacetate decarboxylase-activating enzyme
- Fe/S:
-
Iron–sulfur cluster
- ITC:
-
Isothermal titration calorimetry
- Pfl-AE:
-
Pyruvate formate-lyase-activating enzyme
- RS cluster:
-
SAM binding [4Fe-4S]2+/1+ cluster
- WT-AE:
-
Wild-type 4Hpad-AE (12.0 ± 0.3 mol Fe per mol protein)
References
Yu L, Blaser M, Andrei PI, Pierik AJ, Selmer T (2006) Biochemistry 45:9584–9592
Martins BM, Blaser M, Feliks M, Ullmann GM, Buckel W, Selmer T (2011) J Am Chem Soc 133:14666–14674
D’Ari L, Barker HA (1985) Arch Microbiol 143:311–312
Feliks M, Martins BM, Ullmann GM (2013) J Am Chem Soc 135:14574–14585
Dawson LF, Stabler RA, Wren BW (2008) J Med Microbiol 57:745–749
Shisler K, Broderick JB (2014) Arch Biochem Bioph 546:64–71
Eklund H, Fontecave M (1999) Structure 7:257–262
Selvaraj B, Pierik AJ, Bill E, Martins BM (2013) J Biol Inorg Chem 18:633–643
Frey PA, Hegeman AD, Ruzicka FJ (2008) Crit Rev Biochem Mol Biol 43:63–88
Lanz ND, Booker SJ (2012) Biochim Biophys Acta 1824:1196–1212
Knappe J, Neugebauer FA, Blaschkowski HP, Gänzler M (1984) Proc Natl Acad Sci 81:1332–1335
Gambarelli S, Luttringer F, Padovani D, Mulliez E, Fontecave M (2005) Chem Biochem 6:1960–1962
Selmer T, Pierik AJ, Heider J (2005) Biol Chem 386:981–988
Craciun S, Balskus EP (2012) Proc Natl Acad Sci 109:21307–21312
Demick JM, Lanzilotta WN (2011) Biochemistry (Us) 50:440–442
Fish WW (1988) Methods Enzymol 158:357–364
Sweeney WV, Rabinowitz JC (1980) Ann Rev Biochem 49:139–161
Gütlich P, Bill E, Trautwein AX (2011) Mössbauer spectroscopy and transition metal chemistry. Springer, Berlin
Leatherbarrow RJ (2009) GraFit Version 5.5.0 Ed., Erithacus Software Limited, Horley
Vey JL, Yang J, Li M, Broderick WE, Broderick JB, Drennan CL (2008) Proc Natl Acad Sci 105:16137–16141
Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, Broderick JB (2005) Inorg Chem 44:727–741
Ollagnier S, Mulliez E, Schmidt PP, Eliasson R, Gaillard J, Deronzier C, Bergman T, Graslund A, Reichard P, Fontecave M (1997) J Biol Chem 272:24216–24223
Yang J, Naik SG, Ortillo DO, Garcia-Serres R, Li M, Broderick WE, Huynh BH, Broderick JB (2009) Biochemistry 48:9234–9241
Hilberg M, Pierik AJ, Bill E, Friedrich T, Lippert ML, Heider J (2012) J Biol Inorg Chem 17:49–56
Benjdia A, Subramanian S, Leprince J, Vaudry H, Johnson MK, Berteau O (2010) FEBS J 277:1906–1920
Grove T, Ahlum JH, Qin RM, Lanz ND, Radle MI, Krebs C, Booker SJ (2013) Biochemistry 52:2874–2887
Goldman PJ, Grove TL, Sites LA, McLaughlin MI, Booker SJ, Drennan CL (2013) Proc Natl Acad Sci 110:8519–8524
Grove TL, Lee KH, St Clair J, Krebs C, Booker SJ (2008) Biochemistry 47:7523–7538
Peng Y, Veneziano SE, Gillispie GD, Broderick JB (2010) J Biol Chem 285:27224–27231
Acknowledgments
The Deutsche Forschungsgemeinschaft (DFG) (Grants MA 3348/2-1 and MA 3348/2-2) supported this work. We thank Thorsten Selmer (FH Aachen, Germany) for the 4Hpad clone, Silke Steinborn and Rainer Dietrich (Strukturbiologie/Biochemie) for technical assistance; Bernd Mienert (MPI-CEC) for recording the Mössbauer spectra; and all members of the Strukturbiologie/Biochemie group for helpful discussions, especially Tobias Werther for help in the analysis of the ITC experiments. We are grateful to Wolfgang Buckel (MPI für terrestrische Mikrobiologie, Marburg, Germany) for continuous support and helpful discussions. We also acknowledge the reviewers for their helpful suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Selvaraj, B., Pierik, A.J., Bill, E. et al. The ferredoxin-like domain of the activating enzyme is required for generating a lasting glycyl radical in 4-hydroxyphenylacetate decarboxylase. J Biol Inorg Chem 19, 1317–1326 (2014). https://doi.org/10.1007/s00775-014-1189-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-014-1189-3