Abstract
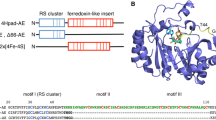
4-Hydroxyphenylacetate decarboxylase (4Hpad) is an Fe/S cluster containing glycyl radical enzyme (GRE), which catalyses the last step of tyrosine fermentation in clostridia, generating the bacteriostatic p-cresol. The respective activating enzyme (4Hpad-AE) displays two cysteine-rich motifs in addition to the classical S-adenosylmethionine (SAM) binding cluster (RS cluster) motif. These additional motifs are also present in other glycyl radical activating enzymes (GR-AE) and it has been postulated that these orthologues may use an alternative SAM homolytic cleavage mechanism, generating a putative 3-amino-3-carboxypropyl radical and 5′-deoxy-5′-(methylthio)adenosine but not a 5′-deoxyadenosyl radical and methionine. 4Hpad-AE produced from a codon-optimized synthetic gene binds a maximum of two [4Fe–4S]2+/+ clusters as revealed by EPR and Mössbauer spectroscopy. The enzyme only catalyses the turnover of SAM under reducing conditions, and the reaction products were identified as 5′-deoxyadenosine (quenched form of 5′-deoxyadenosyl radical) and methionine. We demonstrate that the 5′-deoxyadenosyl radical is the activating agent for 4Hpad through p-cresol formation and correlation between the production of 5′-deoxyadenosine and the generation of glycyl radical in 4Hpad. Therefore, we conclude that 4Hpad-AE catalyses a classical SAM-dependent glycyl radical formation as reported for GR-AE without auxiliary clusters. Our observation casts doubt on the suggestion that GR-AE containing auxiliary clusters catalyse the alternative cleavage reaction detected for glycerol dehydratase activating enzyme.






Similar content being viewed by others
Abbreviations
- 4Hpad:
-
4-Hydroxyphenylacetate decarboxylase
- 4Hpad-AE:
-
4-Hydroxyphenylacetate decarboxylase activating enzyme
- 4Hpad-AE*:
-
4-Hydroxyphenylacetate decarboxylase activating enzyme treated with Fe/S cluster proteins (12.0 ± 0.3 mol Fe per mole of protein)
- Gdh-AE:
-
Glycerol dehydratase activating enzyme
- GR-AE:
-
Glycyl radical activating enzyme
- GRE:
-
Glycyl radical enzyme
- HPLC:
-
High-performance liquid chromatography
- ISC:
-
Fe/S cluster
- ITC:
-
Isothermal titration calorimetry
- Nrd-AE:
-
Glycyl radical activating enzyme for class III ribonucleotide reductase
- Pfl-AE:
-
Glycyl radical activating enzyme for pyruvate formate lyase
- PITC:
-
Phenylisothiocyanate
- RS cluster:
-
S-Adenosylmethionine-binding [4Fe–4S]2+/1+ cluster
- SAM:
-
S-Adenosylmethionine
- TLC:
-
Thin-layer chromatography
References
Frey PA, Hegerman AD, Reed GH (2006) Chem Rev 106:3302–3316
Buckel W (2009) Angew Chem Int Ed 48:6779–6787
Booker SJ (ed) (2012) Biochim Biophys Acta 1824:1151–1306
Kampmeier JA (2010) Biochemistry 49:10770–10772
Broderick JB (2010) Nature 465:877–878
Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, Wang E, Freed J, Krebs C, Ealick SE, Lin H (2010) Nature 465:891–896
Demick JM, Lanzilotta WN (2011) Biochemistry 50:440–442
Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE (2001) Nucleic Acids Res 29:1097–1106
Krebs C, Broderick WE, Henshaw TF, Broderick JB, Huynh BH (2002) J Am Chem Soc 124:912–913
Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, Broderick JB (2005) Inorg Chem 44:727–741
Frey PA, Hegeman AD, Ruzicka FJ (2008) Crit Rev Biochem Mol Biol 43:63–88
Eklund H, Fontecave M (1999) Structure 7:257–262
Buckel W, Zhang J, Friedrich P, Parthasarathy A, Li H, Djurdjevic I, Dobbek H, Martins BM (2012) Biochim Biophys Acta 1824:1278–1290
Yu L, Blaser M, Andrei PI, Pierik AJ, Selmer T (2006) Biochemistry 45:9584–9592
Martins BM, Blaser M, Feliks M, Ullmann GM, Buckel W, Selmer T (2011) J Am Chem Soc 133:14666–14674
Dawson LF, Stabler RA, Wren BW (2008) J Med Microbiol 57:745–749
Smith EA, Macfarlane GT (1997) Microb Ecol 33:180–188
Selmer T, Pierik AJ, Heider J (2005) Biol Chem 386:981–988
Li L, Patterson DP, Fox CC, Lin B, Coshigano PW, Marsh ENG (2009) Biochemistry 48:1284–1292
Hilberg M, Pierik AJ, Bill E, Friedrich T, Lippert ML, Heider J (2012) J Biol Inorg Chem 17:49–56
Jarling R, Sadeghi M, Drozdowska M, Lahme S, Buckel W, Rabus R, Widdel F, Golding BT, Wilkes W (2012) Angew Chem Int Ed 51:1334–1338
Knappe J, Neugebauer FA, Blaschkowski HP, Gänzler M (1984) Proc Natl Acad Sci USA 81:1332–1335
Gambarelli S, Luttringer F, Padovani D, Mulliez E, Fontecave M (2005) Chem Biochem 6:1960–1962
Lanz ND, Booker SJ (2012) Biochim Biophys Acta 1824:1196–1212
Sharp PM, Li W-H (1987) Nucleic Acids Res 15:1281–1295
Nakamura M, Saeki K, Takahashi Y (1999) J Biochem 126:10–18
Fish WW (1988) Methods Enzymol 158:357–364
Sweeney WV, Rabinowitz JC (1980) Annu Rev Biochem 49:139–161
Gütlich P, Bill E, Trautwein AX (2011) Mössbauer spectroscopy and transition metal chemistry. Springer, Berlin
Hui AK, Armstrong BH, Wray AA (1978) J Quant Spectrosc Radiat Transf 19:509–516
Schreier F (2011) J Quant Spectrosc Radiat Transf 112:1010–1025
Dimova N (2003) C R Acad Bulg Sci 56:75–78
Ollagnier S, Mulliez E, Schmidt PP, Eliasson R, Gaillard J, Deronzier C, Bergman T, Graslund A, Reichard P, Fontecave M (1997) J Biol Chem 272:24216–24223
Wong KK, Murray BW, Lewisch SA, Baxter MK, Ridky TW, Ulissidemario L, Kozarich JW (1993) Biochemistry 32:14102–14110
Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD (2009) Nat Chem Biol 5:593–599
Duschene KS, Veneziano SE, Silver SC, Broderick JB (2009) Curr Opin Chem Biol 13:74–83
Dey A, Peng Y, Broderick WE, Hedman B, Hodgson KO, Broderick JB, Solomon EI (2011) J Am Chem Soc 133:18656–18662
Jarrett JT (2003) Curr Opin Chem Biol 7:174–182
Vey JL, Yang J, Li M, Broderick WE, Broderick JB, Drennan CL (2008) Proc Natl Acad Sci USA 105:16137–16141
Hinckley GT, Frey PA (2006) Biochemistry 45:6996
Sauter M, Sawers RG (1990) Mol Microbiol 4:355–363
Yang J, Naik SG, Ortillo DO, Garcia-Serres R, Li M, Broderick WE, Huynh BH, Broderick JB (2009) Biochemistry 48:9234–9241
Thauer RK, Jungermann K, Decker K (1977) Bacteriol Rev 41:100–180
Hoffman JL (1986) Biochemistry 25:4444–4449
Acknowledgments
The Deutsche Forschungsgemeinschaft (grants MA 3348/2-1 and MA 3348/2-2) supported this work. Silke Steinborn and Rainer Dietrich (Strukturbiologie/Biochemie group) are acknowledged for excellent technical assistance. We thank Angelica Woyda (Analytische Chemie und Umweltchemie, Humboldt-Universität zu Berlin) for collecting electrospray ionization mass spectrometry data, Bern Miniert (MPI-CEC) for recording the Mössbauer spectra and Tobias Werther for help in the analysis of the ITC experiments. We are grateful to Thorsten Selmer (Fachhochschule Aachen, Germany) for the 4Hpad clone and all members of the Strukturbiologie/Biochemie group for helpful discussions. We also acknowledge the reviewers for valuable suggestions and Wolfgang Buckel (MPI für Terrestrische Mikrobiologie, Marburg, Germany) for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Selvaraj, B., Pierik, A.J., Bill, E. et al. 4-Hydroxyphenylacetate decarboxylase activating enzyme catalyses a classical S-adenosylmethionine reductive cleavage reaction. J Biol Inorg Chem 18, 633–643 (2013). https://doi.org/10.1007/s00775-013-1008-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-013-1008-2