Abstract
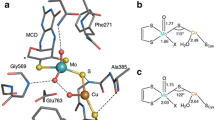
Carbon monoxide dehydrogenase from the bacterium Oligotropha carboxidovorans catalyzes the oxidation of CO to CO2 at a unique [CuSMoO2] cluster. In the bacteria the cluster is assembled post-translational. The integration of S, and particularly of Cu, is rate limiting in vivo, which leads to CO dehydrogenase preparations containing the mature and fully functional enzyme along with forms of the enzyme deficient in one or both of these elements. The active sites of mature and immature forms of CO dehydrogenase were converted into a [MoO3] centre by treatment with potassium cyanide. We have established a method, which rescues 50% of the CO dehydrogenase activity by in vitro reconstitution of the active site through the supply of sulphide first and subsequently of Cu(I) under reducing conditions. Immature forms of CO dehydrogenase isolated from the bacterium, which were deficient in S and/or Cu at the active site, were similarly activated. X-ray crystallography and electron paramagnetic resonance spectroscopy indicated that the [CuSMoO2] cluster was properly reconstructed. However, reconstituted CO dehydrogenase contains mature along with immature forms. The chemical reactions of the reconstitution of CO dehydrogenase are summarized in a model, which assumes resulphuration of the Mo-ion at both equatorial positions at a 1:1 molar ratio. One equatorial Mo–S group reacts with Cu(I) in a productive fashion yielding a mature, functional [CuSMoO2] cluster. The other Mo–S group reacts with Cu(I), then Cu2S is released and an oxo group is introduced from water, yielding an inactive [MoO3] centre.






Similar content being viewed by others
Abbreviations
- AAS:
-
Atomic absorption spectroscopy
- EPR:
-
Electron paramagnetic resonance
- MCD:
-
Molybdopterin cytosine dinucleotide
- OD:
-
optical density
- ORF:
-
open reading frame
- PAGE:
-
polyacrylamide gel electrophoresis
- SDS:
-
sodium dodecyl sulphate
References
Meyer O, Gremer L, Ferner R, Ferner M, Dobbek H, Gnida M, Meyer-Klauke W, Huber R (2000) Biol Chem 381:865–876
Dobbek H, Gremer L, Kiefersauer R, Huber R, Meyer O (2002) Proc Natl Acad Sci USA 99:15971–15976
Gnida M, Ferner R, Gremer L, Meyer O, Meyer-Klaucke W (2003) Biochemistry 42:222–230
Dobbek H, Gremer L, Meyer O, Huber R (1999) Proc Natl Acad Sci USA 96:8884–8889
Gremer L, Kellner S, Dobbek H, Huber R, Meyer O (2000) J Biol Chem 275:1864–1872
Fuhrmann S, Ferner M, Jeffke T, Henne A, Gottschalk G, Meyer O (2003) Gene 322:67–75
Meyer O, Schlegel HG (1978) Arch Microbiol 118:35–43
Meyer O, Stackebrandt E, Auling G (1993) Syst Appl Microbiol 16:390–395
Kraut M, Hugendieck I, Herwig S, Meyer O (1989) Arch Microbiol 152:335–341
Okamoto K, Matsumoto K, Hille R, Eger BT, Pai EF, Nishino T (2004) Proc Natl Acad Sci USA 101:7931–7936
Bradford MM (1976) Analyt Biochem 72:248–254
Beisenherz G, Boltze HJ, Bücher T, Czok R, Garbade KH, Meyer-Arendt E, Pfleiderer G (1953) Z Naturforschg 8:555–577
Meyer O, Rajagopalan KV (1984) J Biol Chem 259:5612–5617
Laemmli UK (1970) Nature 227:680–685
Westley J (1981) Methods Enzymol 77:285–291
Minder W, Stocker E (1936) Z Kristallogr 94:137–142
Okaya Y, Knobler CB (1964) Acta Cryst 17:928–930
Morpurgo L, Rotilio G, Hartmann HJ, Weser U (1984) Biochem J 221:923–925
Kiefersauer R, Than M, Dobbek H, Gremer L, Melero M, Strobl S, Dias JM, Soulimane T, Huber R (2000) J Appl Crystallogr 33:1223–1230
Kabsch W (1993) J Appl Crystallogr 26:795–800
Turk D (1996) In: Bourne PE, Watepaugh K (eds) Meeting of the international union of crystallography macromolecular computing school. Bellingham, WA: International Union of Crystallography
Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Acta Cryst D54:905–921
Laskowski RA, McArthur MW, Moss DS, Thornton JM (1993) J Appl Crystallogr 33:491–497
Wahl RC, Rajagopalan KV (1982) J Biol Chem 257:1354–1359
Maiti NC, Tomita T, Kitagawa T, Okamoto K, Nishino T (2003) J Biol Inorg Chem 8:327–333
DeLano WL (2002) The PyMOL user’s manual. DeLano Scientific, San Carlos
Bray RC, George GN, Lange R, Meyer O (1983) Biochem J 211:687–694
Peariso K, Chohan BS, Carrano CJ, Kirk ML (2003) Inorg Chem 42:6194–6203
Acknowledgements
We thank Dr. Lothar Gremer and Jörg Johannes (Microbiology, University of Bayreuth) for the introduction of Cu–thiourea in the reconstitution of CO dehydrogenase, Dr. Lothar Gremer for providing a sample of CO dehydrogenase, and Dr. Reiner Kiefersauer and Professor Robert Huber (Max-Planck-Institut für Biochemie, Martinsried) for the transformation of crystals. This work was supported by grants from by the Deutsche Forschungsgemeinschaft to Ortwin Meyer (ME 732/8-1) and Holger Dobbek (DO 785/1), the Fonds der Chemischen Industrie (Frankfurt am Main, Germany), and contains instrumentation purchased with funds from the Freistaat Bayern, the Bundesministerium für Bildung und Forschung, and the Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Resch, M., Dobbek, H. & Meyer, O. Structural and functional reconstruction in situ of the [CuSMoO2] active site of carbon monoxide dehydrogenase from the carbon monoxide oxidizing eubacterium Oligotropha carboxidovorans . J Biol Inorg Chem 10, 518–528 (2005). https://doi.org/10.1007/s00775-005-0006-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-005-0006-4