Abstract
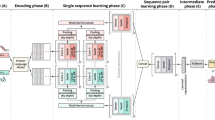
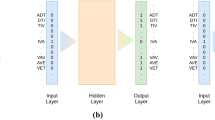
Sequence classification is crucial in predicting the function of newly discovered sequences. In recent years, the prediction of the incremental large-scale and diversity of sequences has heavily relied on the involvement of machine-learning algorithms. To improve prediction accuracy, these algorithms must confront the key challenge of extracting valuable features. In this work, we propose a feature-enhanced protein classification approach, considering the rich generation of multiple sequence alignment algorithms, N-gram probabilistic language model and the deep learning technique. The essence behind the proposed method is that if each group of sequences can be represented by one feature sequence, composed of homologous sites, there should be less loss when the sequence is rebuilt, when a more relevant sequence is added to the group. On the basis of this consideration, the prediction becomes whether a query sequence belonging to a group of sequences can be transferred to calculate the probability that the new feature sequence evolves from the original one. The proposed work focuses on the hierarchical classification of G-protein Coupled Receptors (GPCRs), which begins by extracting the feature sequences from the multiple sequence alignment results of the GPCRs sub-subfamilies. The N-gram model is then applied to construct the input vectors. Finally, these vectors are imported into a convolutional neural network to make a prediction. The experimental results elucidate that the proposed method provides significant performance improvements. The classification error rate of the proposed method is reduced by at least 4.67% (family level I) and 5.75% (family Level II), in comparison with the current state-of-the-art methods. The implementation program of the proposed work is freely available at: https://github.com/alanFchina/CNN.




Similar content being viewed by others
References
Adams MD, Celniker SE, Holt RA et al (2000) The genome sequence of Drosophila melanogaster. Science 287:2185–2195
Altschul SF (1991) Amino acid substitution matrices from an information theoretic perspective. J Mol Biol 219:555–565
Altschul SF, Madden TL, Schaffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Bairoch A, Boeckmann B, Ferro S et al (2004) Swiss-Prot: juggling between evolution and stability. Brief Bioinform 5:39–55
Bandyopadhyay S (2005) An efficient technique for superfamily classification of amino acid sequences: feature extraction, fuzzy clustering and prototype selection. Fuzzy Sets Syst 152(1):5–16
Bhasin M, Raghava GPS (2004) GPCRpred: an SVM-based method for prediction of families and subfamilies of G-protein coupled receptors. Nucl Acids Res 32:383–389
Boeckmann B et al (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL. Nucl Acids Res 31:365–370
Boutet E, Lieberherr D, Tognolli M et al (2016) UniProtKB/Swiss-Prot, the manually annotated section of the UniProt knowledge base: how to use the entry view. Methods Mol Biol 1374:23–54
Brown PF, Desouza PV, Mercer RL et al (1992) Class-based n-gram models of natural language. Comput Linguist 18(4):467–479
Chambers G, Lawrie L, Cash P et al (2000) Proteomics: a new approach to the study of disease. J Pathol 192:280–288
Cheng BYM, Carbonell JG, Klein-Seetharaman J et al (2005) Protein classification based on text document classification techniques. Proteins Struct Funct Bioinform 58(4):955–970
Daugaard M, Rohde M, Jäättelä M (2007) The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett 581(19):3702–3710
Davies MN, Secker A, Halling-Brown M et al (2008) Gpcrtree: online hierarchical classification of GPCR function. BMC Res Notes 1(1):67
Dayhoff MO, Schwartz R, Orcutt BC (1978) A model of evolutionary change in proteins. In: Davidoff MO (ed) Atlas of protein sequence and structure. National Biomedical Research Foundation, Silver Spring (MD), pp 345–352
Dongardive J, Abraham S (2016) Protein sequence classification based on n-gram and k-nearest neighbor algorithm. Comput Intell Data Min 2:163–171
Durbin R, Eddy S, Krogh A et al (1998) Biological sequence analysis. Cambridge University Press, Cambridge
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32(5):1792–1797
Firdaus MA, Razib MO (2009) Analysis of multiple alignment on the performance of classification algorithm for remote protein homology detection. Genes Dev 22(24):3489–3496
George SR, O’Dowd BF, Lee SP (2002) G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat Rev Drug Discov 1(10):808–820
Gether U (2000) Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr Rev 21(1):90–113
Gosele C, Hong L, Kreitler T et al (2000) High-throughput scanning of the rat genome using interspersed repetitive sequence-PCR markers. Genomics 69:287–294
Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89:10915–10919
Henikoff S, Henikoff JG, Alford WJ, Pietrokovski S (1995) Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene 163(2):17–26
Iqbal MJ, Faye I, Said AM et al (2014) Data mining of protein sequences with amino acid position-based feature encoding technique. In: Proceedings of the first international conference on advanced data and information engineering. Singapore, pp 119–126
Isberg V, Vroling B, Rob VDK et al (1998) GPCRDB: An information system for G protein-coupled receptors. Nucl Acid Res 26(1):275–279
Jeong JC, Lin X, Chen X (2011) On position-specific scoring matrix for protein function prediction. IEEE/ACM Trans Comput Biol Bioinform 8(2):308–315
Kalchbrenner N, Grefenstette E, Blunsom PA (2014) A convolutional neural network for modelling sentences. In: Proceedings of the 52nd annual meeting
Kamal NAM, Bakar AA, Zainudin S (2015) Filter-wrapper approach to feature selection of GPCR protein[C]. In: International conference on electrical engineering and informatics. IEEE, pp 693–698
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. In: Proceedings of the 26th annual conference on neural information processing systems, NIPS 2012, vol 2, pp 1097–1105
Li Z, Zhou X, Dai Z et al (2010) Classification of G-protein coupled receptors based on support vector machine with maximum relevance minimum redundancy and genetic algorithm. BMC Bioinform 11(1):325
Li M, Ling C, Gao J (2017) An efficient CNN-based classification on G-protein coupled receptors using TF-IDF and N-gram. In: Proceedings of 2017 IEEE symposium on computers and communications (ISCC), Heraklion, pp 924–931
Lynch M (2002) Intron evolution as a population-genetic process. Proc Natl Acad Sci USA 99:6118–6123
Mardia KV, Kent JT, Bibby JM (1979) Multivariate analysis. Academic Press, London, p 521
Naveed M, Khan AU (2012) GPCR-MPredictor: multi-level prediction of G protein-coupled receptors using genetic ensemble. Amino Acids 42:1809–1823
Notredame C, Higgins DG, Heringa J (1996) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302(1):205–217
Pearson WR (1996) Effective protein sequence comparison. Methods Enzymol 266:227–258
Pearson WR (1998) Empirical statistical estimates for sequence similarity searches. J Mol Biol 276:71–84
Ramos J (2003) Using tf-idf to determine word relevance in document queries. In: Proceedings of the 1st instructional conference on machine learning
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. Comput Sci
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22(22):4673–4680
Thornton JM (2001) From genome to function. Science 292:2095–2097
Vinga S, Almeida J (2003) Alignment-free sequence comparison: a review. Bioinformatics 19(4):513–523
Vries JK, Munshi R, Tobi D et al (2004) A sequence alignment-independent method for protein classification. Appl Bioinform 3(2):137
Waterston RH, Lindblad-Toh K, Birney E et al (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562
Wu CH, Huang H, Yeh LL et al (2003) Protein family classification and functional annotation. Comput Biol Chem 27:37–47
Yao X (1999) Evolving artificial neural networks. Proc IEEE 87(9):1423–1447
Zavaljevski N, Stevens FJ, Reifman J (2002) Support vector machines with selective kernel scaling for protein classification and identification of key amino acid positions. Bioinformatics 18(5):689–696
Zhang YX, Perry K, Vinci VA et al (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644–646
Acknowledgements
Funding was provided by National Natural Science Foundation of China (Grant no. 61602026).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Research involving human participants and/or animals
The authors did not use human participants or animals in the investigation.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Handling Editor: I. Greger.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, M., Ling, C., Xu, Q. et al. Classification of G-protein coupled receptors based on a rich generation of convolutional neural network, N-gram transformation and multiple sequence alignments. Amino Acids 50, 255–266 (2018). https://doi.org/10.1007/s00726-017-2512-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-017-2512-4