Abstract
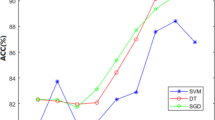
Knowledge of the types of membrane protein provides useful clues in deducing the functions of uncharacterized membrane proteins. An automatic method for efficiently identifying uncharacterized proteins is thus highly desirable. In this work, we have developed a novel method for predicting membrane protein types by exploiting the discrimination capability of the difference in amino acid composition at the N and C terminus through split amino acid composition (SAAC). We also show that the ensemble classification can better exploit this discriminating capability of SAAC. In this study, membrane protein types are classified using three feature extraction and several classification strategies. An ensemble classifier Mem-EnsSAAC is then developed using the best feature extraction strategy. Pseudo amino acid (PseAA) composition, discrete wavelet analysis (DWT), SAAC, and a hybrid model are employed for feature extraction. The nearest neighbor, probabilistic neural network, support vector machine, random forest, and Adaboost are used as individual classifiers. The predicted results of the individual learners are combined using genetic algorithm to form an ensemble classifier, Mem-EnsSAAC yielding an accuracy of 92.4 and 92.2% for the Jackknife and independent dataset test, respectively. Performance measures such as MCC, sensitivity, specificity, F-measure, and Q-statistics show that SAAC-based prediction yields significantly higher performance compared to PseAA- and DWT-based systems, and is also the best reported so far. The proposed Mem-EnsSAAC is able to predict the membrane protein types with high accuracy and consequently, can be very helpful in drug discovery. It can be accessed at http://111.68.99.218/membrane.






Similar content being viewed by others
References
Breiman L (2001) Random forests. Mach Learn 45:5–32
Cai YD, Zhou GP, Chou KC (2003) Support vector machines for predicting membrane protein types by using functional domain composition. Biophys J 84:3257–3263
Cai YD, Ricardo PW, Jen CH, Chou KC (2004) Application of SVM to predict membrane protein types. J Theor Biol 226:373–376
Call ME, Wucherpfennig KW, Chou JJ (2010) The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat Immunol 11:1023–1029
Chou KC (1995) A novel approach to predicting protein structural classes in a (20–1)-D amino acid composition space. Proteins Struct Funct Genet 21:319–344
Chou KC (2001) Prediction of protein subcellular attributes using pseudo-amino acid composition. Proteins Struct Funct Genet 43:246–255
Chou KC, Cai YD (2002) Using functional domain composition and support vector machines for prediction of protein subcellular location. J Biol Chem 277:45765–45769
Chou KC, Cai YD (2005a) Using GO-PseAA predictor to indentify membrane proteins and their types. Biochem Biophys Res Commun 327:845–847
Chou KC, Cai YD (2005b) Prediction of membrane protein types by incorporating amphipathic effects. J Chem Inf Model 45:407–413
Chou KC, Elrod DW (1999) Prediction of membrane protein types and subcellular locations. Proteins Struct Funct Genet 34:137–153
Chou KC, Shen HB (2006a) Predicting eukaryotic protein subcellular location by fusing optimized evidence-theoretic K-nearest neighbor classifiers. J Proteome Res 5:1888–1897
Chou KC, Shen HB (2006b) Hum-PLoc: a novel ensemble classifier for predicting human protein Subcellular localization. Biochem Biophys Res Commun 347:150–157
Chou KC, Shen HB (2007a) Memtype-2L: a web server for predicting membrane proteins and their types by incorporating evolution information through Pse-PSSM. Biochem Biophys Res Commun 360:339–345
Chou KC, Shen HB (2007b) Euk-mPLoc: a fusion classifier for large-scale eukaryotic protein subcellular location prediction by incorporating multiple sites. J Proteome Res 6:1728–1734
Chou KC, Shen HB (2007c) Review: recent progresses in protein subcellular location prediction. Anal Biochem 370:1–16
Chou KC, Shen HB (2009a) Review: recent advances in developing web-servers for predicting protein attributes. Nat Sci 2:63–92. http://www.scirp.org/journal/NS/
Chou KC, Shen HB (2010a) Cell-PLoc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat Sci 2:1090–1103. http://www.scirp.org/journal/NS/
Chou KC, Shen HB (2010b) A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLoS ONE 5:e9931
Chou JJ, Zhang CT (1993) A joint prediction of the folding types of 1490 human proteins from their genetic codons. J Theor Biol 161:251–262
Chou KC, Zhang CT (1994) Predicting protein folding types by distance functions that make allowances for amino acid interactions. J Biol Chem 269:22014–22020
Khan A, Tahir SF, Majid A, Choi Tae-Sun. (2008a) Machine learning based adaptive watermark decoding in view of an anticipated attack. Pattern Recognit 41:2594–2610
Khan A, Tahir SF, Choi TS (2008b) Intelligent extraction of a digital watermark from a distorted image. IEICE Trans Inf Syst. E91-D 7:2072–2075
Khan A, Khan FM, Choi TS (2008c) Proximity based GPCRs prediction in transform domain. Biochem Biophys Res Commun 371:411–415
Khan A, Majid A, Choi TS (2010) Predicting protein subcellular location: exploiting amino acid based sequence of feature spaces and fusion of diverse classifiers. Amino Acids 38:347–350
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Liu H, Wang M, Chou KC (2005) Low-frequency Fourier spectrum for predicting membrane protein types. Biochem Biophys Res Commun 336:737–739
Mahdavi A, Jahandideh S (2011) Application of density similarities to predict membrane protein types based on pseudo amino acid composition. J Theor Biol 276:132–137
Nakashima H, Nishikawa AO (1986) The folding type of a protein is relevant to the amino acid composition. J Biochem 99:152–162
Nanni L, Lumini A (2006) Ensemblator: an ensemble of classifiers for reliable classification of biological data. Pattern Recognit Lett 28:622–630
Nanni L, Lumini A (2008a) Genetic programming for creating Chou’s pseudo amino acid based features for submitochondria localization. Amino Acids 34:653–660
Nanni L, Lumini A (2008b) An ensemble of support vector machines for predicting the membrane proteins type directly from the amino acid sequences. Amino Acids 35(3):573–580
Nanni L, Brahnam S, Lumini A (2010) High performance set of PseAAC and sequence based descriptors for protein classification. J Theor Biol 266(1):1–10
Pielak RM, Chou JJ (2010) Flu channel drug resistance: a tale of two sites. Protein Cell 1:246–258
Qiu JD, Sun XU, Huang JH, Liang RP (2010) Prediction of the types of membrane proteins based on discrete wavelet transform and support vector machines. Protien J 29:114–119
Rezaei MA, Maleki PA, Karami Z, Asadabadi EB, Sherafat MA, Moghaddam KA, Fadaie M, Forouzanfar M (2008) Prediction of membrane protein types by means of wavelet analysis and cascaded neural network. J Theor Biol 255:817–820
Schapire RE, Freund Y, Bartlett P, Lee WS (1998) Boosting the margin a new explanation for the effectiveness of voting methods. Ann Stat 26:1651–1686
Shen HB, Chou KC (2007) Using ensemble classifier to identify membrane protein types. Amino Acids 32:483–488
Tsoumakas G, Katakis I (2007) Multi-label classification: an overview. Int J Data Wareh Min 3:1–13
Wang M, Yang J, Liu GP, Xu ZJ, Chou KC (2004) Weighted-support vector machines for predicting membrane protein types based on pseudo amino acid composition. Protein Eng Des Sel 17:509–516
Wens Z, Wang K, Li M, Nie F (2005) Analyzing functional similarity of protein sequences with discrete wavelet transform. Comput Biol Chem 29:220–228
Zhang CX, Zhang JS (2008) RotBoost: a technique for combining Rotation Forest and AdaBoost. Pattern Recognit Lett. doi:10.1016/j.patrec.2008.03.006
Zhou XB, Chen C, Li ZC, Zou XY (2007) Using Chou’s amphiphilic pseudo-amino acid composition and support vector machine for prediction of enzyme subfamily classes. J Theor Biol 248:546–551
Acknowledgments
This work is supported by the Higher Education Commission of Pakistan under the indigenous PhD scholarship program 17-5-3 (Eg3-045)/HEC/Sch/2006).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hayat, M., Khan, A. & Yeasin, M. Prediction of membrane proteins using split amino acid and ensemble classification. Amino Acids 42, 2447–2460 (2012). https://doi.org/10.1007/s00726-011-1053-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-011-1053-5