Abstract
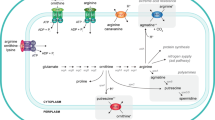
The structure and function of a cadaverine–lysine antiporter CadB and a putrescine–ornithine antiporter PotE in Escherichia coli were evaluated using model structures based on the crystal structure of AdiC, an agmatine–arginine antiporter, and the activities of various CadB and PotE mutants. The central cavity of CadB, containing the substrate binding site, was wider than that of PotE, mirroring the different sizes of cadaverine and putrescine. The size of the central cavity of CadB and PotE was dependent on the angle of transmembrane helix 6 (TM6) against the periplasm. Tyr73, Tyr89, Tyr90, Glu204, Tyr235, Asp303, and Tyr423 of CadB, and Cys62, Trp201, Glu207, Trp292, and Tyr425 of PotE were strongly involved in the antiport activities. In addition, Trp43, Tyr57, Tyr107, Tyr366, and Tyr368 of CadB were involved preferentially in cadaverine uptake at neutral pH, while only Tyr90 of PotE was involved preferentially in putrescine uptake. The results indicate that the central cavity of CadB consists of TMs 2, 3, 6, 7, 8, and 10, and that of PotE consists of TMs 2, 3, 6, and 8. These results also suggest that several amino acid residues are necessary for recognition of cadaverine in the periplasm because the level of cadaverine is much lower than that of putrescine in the periplasm at neutral pH. All the amino acid residues identified as being strongly involved in both the antiport and uptake activities were located on the surface of the transport path consisting of the central cavity and TM12.






Similar content being viewed by others
Abbreviations
- AdiC:
-
Agmatine–arginine antiporter
- CadB:
-
Cadaverine–lysine antiporter
- PotE:
-
Putrescine–ornithine antiporter
- APC superfamily:
-
Amino acid/polyamine/organocation superfamily
- TM:
-
Transmembrane helix
References
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4:1–13
Fang Y, Jayaram H, Shane T, Kolmakova-Partensky L, Wu F, Williams C, Xiong Y, Miller C (2009) Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature 460:1040–1043
Gao X, Lu F, Zhou L, Dang S, Sun L, Li X, Wang J, Shi Y (2009) Structure and mechanism of an amino acid antiporter. Science 324:1565–1568
Gao X, Zhou L, Jiao X, Lu F, Yan C, Zeng X, Wang J, Shi Y (2010) Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 463:828–832
Gitlitz PH, Sunderman FW Jr, Hohnadel DC (1974) Ion-exchange chromatography of amino acids in sweat collected from healthy subjects during sauna bathing. Clin Chem 20:1305–1312
Gong S, Richard H, Foster JW (2003) YjdE (AdiC) is the arginine: agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J Bacteriol 185:4402–4409
Herbert JD, Coulson RA, Hernandez T (1966) Free amino acids in the caiman and rat. Comp Biochem Physiol 17:583–598
Higashi K, Ishigure H, Demizu R, Uemura T, Nishino K, Yamaguchi A, Kashiwagi K, Igarashi K (2008) Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J Bacteriol 190:872–878
Igarashi K, Kashiwagi K (1996) Polyamine transport in Escherichia coli. Amino Acids 10:83–97
Igarashi K, Kashiwagi K (1999) Polyamine transport in bacteria and yeast. Biochem J 344:633–642
Igarashi K, Kashiwagi K (2010a) Characteristics of cellular polyamine transport in prokaryotes and eukaryotes. Plant Physiol Biochem 48:506–512
Igarashi K, Kashiwagi K (2010b) Modulation of cellular function by polyamines. Int J Biochem Cell Biol 42:39–51
Igarashi K, Kashiwagi K, Hamasaki H, Miura A, Kakegawa T, Hirose S, Matsuzaki S (1986) Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J Bacteriol 166:128–134
Iyer R, Williams C, Miller C (2003) Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol 185:6556–6561
Jack DL, Paulsen IT, Saier MH Jr (2000) The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146:1797–1814
Kashiwagi K, Igarashi K (1988) Adjustment of polyamine contents in Escherichia coli. J Bacteriol 170:3131–3135
Kashiwagi K, Hosokawa N, Furuchi T, Kobayashi H, Sasakawa C, Yoshikawa M, Igarashi K (1990) Isolation of polyamine transport-deficient mutants of Escherichia coli and cloning of the genes for polyamine transport proteins. J Biol Chem 265:20893–20897
Kashiwagi K, Suzuki T, Suzuki F, Furuchi T, Kobayashi H, Igarashi K (1991) Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem 266:20922–20927
Kashiwagi K, Miyamoto S, Suzuki F, Kobayashi H, Igarashi K (1992) Excretion of putrescine by the putrescine–ornithine antiporter encoded by the potE gene of Escherichia coli. Proc Natl Acad Sci U S A 89:4529–4533
Kashiwagi K, Shibuya S, Tomitori H, Kuraishi A, Igarashi K (1997) Excretion and uptake of putrescine by the PotE protein in Escherichia coli. J Biol Chem 272:6318–6323
Kashiwagi K, Kuraishi A, Tomitori H, Igarashi A, Nishimura K, Shirahata A, Igarashi K (2000) Identification of the putrescine recognition site on polyamine transport protein PotE. J Biol Chem 275:36007–36012
Kurihara S, Tsuboi Y, Oda S, Kim HG, Kumagai H, Suzuki H (2009) The putrescine importer PuuP of Escherichia coli K-12. J Bacteriol 191:2776–2782
Meng SY, Bennett GN (1992) Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J Bacteriol 174:2659–2669
Miyamoto S, Kashiwagi K, Ito K, Watanabe S, Igarashi K (1993) Estimation of polyamine distribution and polyamine stimulation of protein synthesis in Escherichia coli. Arch Biochem Biophys 300:63–68
Soksawatmaekhin W, Kuraishi A, Sakata K, Kashiwagi K, Igarashi K (2004) Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol Microbiol 51:1401–1412
Soksawatmaekhin W, Uemura T, Fukiwake N, Kashiwagi K, Igarashi K (2006) Identification of the cadaverine recognition site on the cadaverine–lysine antiporter CadB. J Biol Chem 281:29213–29220
Takayama M, Ohyama T, Igarashi K, Kobayashi H (1994) Escherichia coli cad operon functions as a supplier of carbon dioxide. Mol Microbiol 11:913–918
Watson N, Dunyak DS, Rosey EL, Slonczewski JL, Olson ER (1992) Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol 174:530–540
Acknowledgments
We thank Drs. A. J. Michael and K. Williams for their help in preparing this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tomitori, H., Kashiwagi, K. & Igarashi, K. Structure and function of polyamine-amino acid antiporters CadB and PotE in Escherichia coli . Amino Acids 42, 733–740 (2012). https://doi.org/10.1007/s00726-011-0989-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-011-0989-9