Abstract
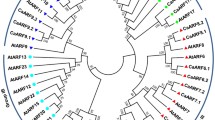
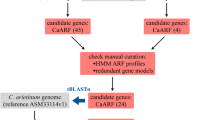
Auxin response factor (ARF) is an important transcription factor that regulates the expression of auxin-responsive genes by direct binding to their promoters, which play a central role in plant growth, development, and response to abiotic stresses. The availability of the entire Coix (Coix lacryma-jobi L.) genome sequence provides an opportunity to investigate the characteristics and evolutionary history of the ARF gene family in this medicine and food homology plant for the first time. In this study, a total of 27 ClARF genes were identified based on the genome-wide sequence of Coix. Twenty-four of the 27 ClARF genes were unevenly distributed on 8 chromosomes except Chr 4 and 10, and the remaining three genes (ClARF25-27) were not assigned to any chromosome. Most of the ClARF proteins were predicted to be localized to the nucleus, except ClARF24, which was localized to both the plasma membrane and nucleus. Twenty-seven ClARFs were clustered into six subgroups based on the phylogenetic analysis. Duplication analysis showed that segmental duplication, rather than tandem duplications promoting the expansion of the ClARF gene family. Synteny analysis showed that purifying selection might have been a primary driving force in the development of the ARF gene family in Coix and other investigated cereal plants. The prediction of the cis element of the promoter showed that 27 ClARF genes contain several stress response elements, suggesting that ClARFs might be involved in the abiotic stress response. Expression profile analysis shows that 27 ClARF genes were all expressed in the root, shoot, leaf, kernel, glume, and male flower of Coix with varying expression levels. Furthermore, qRT-PCR analyses revealed that the majority of ClARFs members were upregulated or downregulated in response to hormone treatment and abiotic stress. The current study expands our understanding of the functional roles of ClARFs in stress responses and provides basic information for the ClARF genes.









Similar content being viewed by others
Data availability
All data and materials used in this research are publicly available. Other supporting data are provided as supplementary files with the manuscript.
Abbreviations
- ARF:
-
Auxin response factor
- DBD:
-
DNA-binding domain
- AuxREs:
-
Auxin-responsive elements
- qRT-PCR:
-
Reverse transcription-quantitative PCR
- Aux/IAA:
-
Auxin/indole-3-acetic acid
- MW:
-
Molecular weight
- PI:
-
Isoelectric point
- ABA:
-
Abscisic acid
- SA:
-
Salicylic acid
References
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408(6814):796–815
Attia KA, Abdelkhalik AF, Ammar MH et al (2009) Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Curr Issues Mol Biol 11(Suppl 1):i29-34
Bouzroud S, Gouiaa S, Hu N et al (2018) Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS One 13(2):e0193517
Cannon SB, Mitra A, Baumgarten A et al (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Chandler JW (2016) Auxin response factors. Plant Cell Environ 39(5):1014–1028
Chen D, Wang W, Wu Y et al (2019) Expression and distribution of the auxin response factors in Sorghum bicolor during development and temperature stress. Int J Mol Sci 20(19):4816
Chen C, Chen H, Zhang Y et al (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202
Chenna R, Sugawara H, Koike T et al (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31(13):3497–3500
Cui J, Li X, Li J et al (2020) Genome-wide sequence identification and expression analysis of ARF family in sugar beet (Beta vulgaris L.) under salinity stresses. PeerJ 8:e9131
Du H, Liu H, Xiong L (2013) Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front Plant Sci 4:397
Fang T, Jiang YX, Chen L et al (2020) Coix seed oil exerts an anti-triple-negative breast cancer effect by disrupting miR-205/S1PR1 axis. Front Pharmacol 11:529962
Goff SA, Ricke D, Lan TH et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296(5565):92–100
Guo C, Wang Y, Yang A et al (2020) The Coix genome provides insights into Panicoideae evolution and papery hull domestication. Mol Plant 13(2):309–320
Ha CV, Le DT, Nishiyama R et al (2013) The auxin response factor transcription factor family in soybean: genome-wide identification and expression analyses during development and water stress. DNA Res 20(5):511–524
Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49(3–4):373–385
Jain M, Khurana JP (2009) Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J 276(11):3148–3162
Jin L, Yarra R, Zhou L et al (2022) The auxin response factor (ARF) gene family in Oil palm (Elaeis guineensis Jacq.): genome-wide identification and their expression profiling under abiotic stresses. Protoplasma 259(1):47–60
Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci U S A 94(22):11786–11791
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Kuo CC, Chen HH,Chiang W (2012) Adlay (yi yi; “soft-shelled job’s tears”; the seeds of Coix lachryma- jobi L. var. ma-yuen Stapf) is a potential cancer chemopreventive agent toward multistage carcinogenesis processes. J Tradit Complement Med 2(4):267–275
Lescot M, Dehais P, Thijs G et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Li XK, Gu K, Liang MW et al (2020) Research progress on chemical constituents and pharmacological effects of Coicis Semen. Chin Tradit Her Drugs 51:5045–5657
Liu Y, Jiang HY, Chen WJ (2011) Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul 63:225–234
Liu N, Dong L, Deng X et al (2018) Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L. BMC Plant Biol 18(1):336
Liu H, Shi J, Cai Z et al (2020) Evolution and domestication footprints uncovered from the genomes of Coix. Mol Plant 13(2):295–308
Mascher M, Gundlach H, Himmelbach A et al (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544(7651):427–433
Okushima Y, Overvoorde PJ, Arima K et al (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17(2):444–463
Okushima Y, Fukaki H, Onoda M et al (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19(1):118–130
Paterson AH, Bowers JE, Bruggmann R et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457(7229):551–556
Remington DL, Vision TJ, Guilfoyle TJ et al (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135(3):1738–1752
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115
Su L, Xu M, Zhang J et al (2021) Genome-wide identification of auxin response factor (ARF) family in kiwifruit (Actinidia chinensis) and analysis of their inducible involvements in abiotic stresses. Physiol Mol Biol Plants 27(6):1261–1276
Tang Y, Bao X, Liu K et al (2018) Genome-wide identification and expression profiling of the auxin response factor (ARF) gene family in physic nut. PLoS ONE 13(8):e0201024
Tombuloglu H (2019) Genome-wide analysis of the auxin response factors (ARF) gene family in barley (Hordeum vulgare L.). J Plant Biochem Biotechnol 28:14–24
Ulmasov T, Hagen G, Guilfoyle TJ (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276(5320):1865–1868
Waadt R, Seller CA, Hsu PK et al (2022) Plant hormone regulation of abiotic stress responses. Nat Rev Mol Cell Biol 23(10):680–694
Wang D, Pei K, Fu Y et al (2007) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394(1–2):13–24
Wang S, Bai Y, Shen C et al (2010) Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomics 10(4):533–546
Wang L, Chen C, Su A et al (2016) Structural characterization of phenolic compounds and antioxidant activity of the phenolic-rich fraction from defatted adlay (Coix lachryma-jobi L. var. ma-yuen Stapf) seed meal. Food Chem 196:509–517
Wang XX, Han YY, Shi GY (2020) Analysis and monitoring of traditional Chinese medicines for tumor based on data mining. Zhongguo Zhong Yao Za Zhi 45(2):444–450
Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot 95(5):707–735
Wu B, Wang L, Pan G et al (2020) Genome-wide characterization and expression analysis of the auxin response factor (ARF) gene family during melon (Cucumis melo L.) fruit development. Protoplasma 257(3):979–992
Xing H, Pudake RN, Guo G et al (2011) Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12:178
Zeng Y, Yang J, Chen J et al (2022) Actional mechanisms of active ingredients in functional food Adlay for human health. Molecules 27(15):4808
Funding
This research work is financially supported by Key project at central government level: The ability to establish sustainable use for valuable Chinese medicine resources (2060302), the Zhejiang Key Scientific and Technological Grant to Breed New Agricultural Varieties (2021C02074), and the Zhejiang Sci-Tech University scientific research fund (19042142-Y).
Author information
Authors and Affiliations
Contributions
YZ, XS, YS, and DW conceived and designed the experiments. YZ, QL, and NM performed the experiments; YZ, XZ, QJ, and ZL validated the data; QJ and ZS revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Handling Editor: Bhumi Nath Tripathi
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhai, Y., Shen, X., Sun, Y. et al. Genome-wide investigation of ARF transcription factor gene family and its responses to abiotic stress in Coix (Coix lacryma-jobi L.). Protoplasma 260, 1389–1405 (2023). https://doi.org/10.1007/s00709-023-01855-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-023-01855-5