Abstract
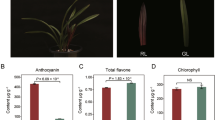
Plantagos are important economical and medicinal plants that possess several bioactive secondary metabolites, such as phenolics, iridoids, triterpenes, and alkaloids. Triterpenoids are the ubiquitous and dynamic secondary metabolites that are deployed by plants for chemical interactions and protection under biotic/abiotic stress. Plantago ovata, a cultivated species, is the source of psyllium, while Plantago major, a wild species, has significant therapeutic potential. Wild species are considered more tolerant to stressful conditions in comparison to their cultivated allies. In view of this, the present study aimed to decipher the terpenoid biosynthetic pathway operative in P. ovata and P. major using a comparative transcriptomics approach. Majority of terpenoid biosynthetic genes were observed as upregulated in P. major including rate limiting genes of MVA (HMGR) and MEP (DXR) pathways and genes (α-AS, BAS, SM, and CYP716) involved in ursolic acid biosynthesis, an important triterpenoid prevalent in Plantago species. The HPLC output further confirmed the higher concentration of ursolic acid in P. major as compared to P. ovata leaf samples, respectively. In addition to terpenoid biosynthesis, KEGG annotation revealed the involvement of differentially expressed unigenes in several metabolic pathways, aminoacyl-tRNA biosynthesis, biosynthesis of antibiotics, and biosynthesis of secondary metabolites. MYB was found as the most abundant transcription factor family in Plantago transcriptome. We have been able to generate valuable information which can help in improving terpenoid production in Plantago. Additionally, the present study has laid a strong foundation for deciphering other important metabolic pathways in Plantago.






Similar content being viewed by others
Data availability
The Illumina sequence data have been submitted to NCBI sequence read archive (SRA) under accession number PRJNA687882.
Code availability
Not applicable.
References
Abdel-Salam EM, Faisal M, Alatar AA et al (2020) Comparative analysis between wild and cultivated cucumbers reveals transcriptional changes during domestication process. Plants 9(1):63
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aminfar Z, Rabiei B, Tohidfar M et al (2019) Identification of key genes involved in the biosynthesis of triterpenic acids in the mint family. Sci Rep 9(1):1–15
Asaf S, Khan AL, Khan A et al (2020) Expanded inverted repeat region with large scale inversion in the first complete plastid genome sequence of Plantago ovata. Sci Rep 10(1):1–16
Bansal S, Narnoliya LK, Mishra B et al (2018) HMG-CoA reductase from camphor tulsi (Ocimum kilimandscharicum) regulated MVA dependent biosynthesis of diverse terpenoids in homologous and heterologous plant systems. Sci Rep 8(1):1–15
Batyrshina ZS, Yaakov B, Shavit R, Singh A, Tzin V (2020) Comparative transcriptomic and metabolic analysis of wild and domesticated wheat genotypes reveals differences in chemical and physical defense responses against aphids. BMC Plant Biol 20:19
Bergman ME, Davis B, Phillips MA (2019) Medically useful plant terpenoids: biosynthesis, occurrence, and mechanism of action. Molecules 24(21):3961
Bleeker PM, Mirabella R, Diergaarde PJ et al (2012) Improved herbivore resistance in cultivated tomato with the sesquiterpene biosynthetic pathway from a wild relative. PNAS 109(49):20124–20129
Chen C, Zheng Y, Zhong Y et al (2018) Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genomics 19(1):550
Choi J, Abbai R, Kim Y et al (2017) Molecular characterization of MYB transcription factor genes from Panax ginseng. Russ J Plant Physiol 64(3):398–409
Dhar MK, Kaul S, Sharma P, Gupta M (2011) Plantago ovata: cultivation, genomics, chemistry and therapeutic applications. In: Singh RJ, editor. Genetic resources, chromosome engineering and crop improvement. CRC Press, New York
Ding ZS, Sun XF, Huang SH et al (2015) Response of photosynthesis to short-term drought stress in rice seedlings overexpressing C-4 phosphoenolpyruvate carboxylase from maize and millet. Photosynthetica 53(4):481–488
Do Amaral MN, Arge LWP, Benitez LC et al (2016) Differential expression of photosynthesis-related genes and quantification of gas exchange in rice plants under abiotic stress. Acta Physiol Plant 38(6):153
Dudareva N, Andersson S, Orlova I et al (2005) The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc Natl Acad Sci 102(3):933–938
Fan H, Li K, Yao F et al (2019) Comparative transcriptome analyses on terpenoids metabolism in field-and mountain-cultivated ginseng roots. BMC Plant Biol 19(1):82
Galvez M, Martın-Cordero C, Lopez-Lazaro M et al (2003) Cytotoxic effect of Plantago spp. on cancer cell lines. J Ethnopharmacol 88(2–3):125–130
Ghasemi S, Kumleh HH, Kordrostami M (2019) Changes in the expression of some genes involved in the biosynthesis of secondary metabolites in Cuminum cyminum L under UV stress. Protoplasma 256(1):279–290
Götz S, García-Gómez JM, Terol J et al (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acid Res 36(10):3420–3435
Grabherr MG, Haas BJ, Yassour M et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644
Ho TT, Murthy HN, Park SY (2020) Methyl jasmonate induced oxidative stress and accumulation of secondary metabolites in plant cell and organ cultures. Int J Mol Sci 21(3):716
Hong Y, Wang Z, Shi H et al (2020) Reciprocal regulation between nicotinamide adenine dinucleotide metabolism and abscisic acid and stress response pathways in Arabidopsis. PLoS Genet 16(6):e1008892
Hussan F, Mansor AS, Hassan SN et al (2015) Anti-inflammatory property of Plantago major leaf extract reduces the inflammatory reaction in experimental acetaminophen-induced liver injury. Evid-Based Compl Alt Med 347861. https://doi.org/10.1155/2015/347861
Isah T (2019) Stress and defense responses in plant secondary metabolites production. Biol Res 52(1):39
Jadaun JS, Sangwan NS, Narnoliya LK et al (2017) Over-expression of DXS gene enhances terpenoidal secondary metabolite accumulation in rose-scented geranium and Withania somnifera: active involvement of plastid isoprenogenic pathway in their biosynthesis. Physiol Plant 159(4):381–400
Jamilah J, Sharifa A, Sharifah N (2012) GC-MS analysis of various extracts from leaf of Plantago major used as traditional medicine. World Appl Sci J 17:67–70
Kartini SP, Thongpraditchote S, Siripong P (2017) Effects of Plantago major extracts and its chemical compounds on proliferation of cancer cells and cytokines production of lipopolysaccharide-activated THP-1 macrophages. Pharmacogn Mag 13(51):393
Koley S, Grafahrend-Belau E, Raorane ML et al (2020) The mevalonate pathway contributes to monoterpene production in peppermint. bioRxiv. https://doi.org/10.1101/2020.05.29.124016
Kotwal S, Kaul S, Sharma P et al (2016) De novo transcriptome analysis of medicinally important Plantago ovata using RNA-Seq. PLoS ONE 11(3):723–729
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Meth 9(4):357
Li R, Chen P, Zhu L et al (2021) Characterization and function of the 1-deoxy-D-xylose-5-phosphate synthase (DXS) gene related to terpenoid synthesis in Pinus massoniana. Int J Mol Sci 22:848. https://doi.org/10.3390/ijms22020848
Li S, Wang H, Li F (2015) The maize transcription factor EREB 58 mediates the jasmonate-induced production of sesquiterpene volatiles. The Plant J 84(2):296–308
Lindgreen S (2012) AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes 5(1):337
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Long M, Shou J, Wang J et al (2020) Ursolic acid limits salt-induced oxidative damage by interfering with nitric oxide production and oxidative defense machinery in rice. Front Plant Sci 11:697
Matsui A, Ishida J, Morosawa T et al (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49(8):1135–1149
Mendoza-Poudereux I, Kutzner E, Huber C, Segura J et al (2015) Metabolic cross-talk between pathways of terpenoid backbone biosynthesis in spike lavender. Plant Physiol Biochem 95:113–120
Meraj TA, Fu J, Raza MA et al (2020) Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes (Basel) 11(4):631–635
Mujeeb F, Bajpai P, Pathak N (2014) Phytochemical evaluation, antimicrobial activity, and determination of bioactive components from leaves of Aegle marmelos. BioMed Res Int. https://doi.org/10.1155/2014/497606
Najafian Y, Hamedi SS, Farshchi MK et al (2018) Plantago major in traditional Persian medicine and modern phytotherapy: a narrative review. Electron Physician 10(2):6390
Nasrollahi V, Mirzaie-asl A, Piri K et al (2014) The effect of drought stress on the expression of key genes involved in the biosynthesis of triterpenoid saponins in liquorice (Glycyrrhiza glabra). Phytochemistry 103:32–37
Nazarizadeh A, Mikaili P, Moloudizargari M et al (2013) Therapeutic uses and pharmacological properties of Plantago major L. and its active constituents. J Basic Appl Sci Res 3(9):212–21
Nkembo MK, Lee JB, Nakagiri T, Hayashi T (2006) Involvement of 2-C-methyl-D-erythritol-4-phosphate pathway in biosynthesis of aphidicolin-like tetracyclic diterpene of Scoparia dulcis. Chem Pharm Bull 54(5):758–760
Nieuwenhuizen NJ, Chen X, Wang MY et al (2015) Natural variation in monoterpene synthesis in kiwifruit: transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiol 167(4):1243–1258
Opitz S, Nes WD, Gershenzon J (2014) Both methylerythritol phosphate and mevalonate pathways contribute to biosynthesis of each of the major isoprenoid classes in young cotton seedlings. Phytochem 98:110–119
Rathinam M, Mishra P, Vasudevan M, Budhwar R et al (2019) Comparative transcriptome analysis of pigeonpea, Cajanus cajan (L.) and one of its wild relatives Cajanus platycarpus (Benth.) Maesen. PloS one 14:e0218731
Rather GA, Sharma A, Jeelani SM et al (2019) Metabolic and transcriptional analyses in response to potent inhibitors establish MEP pathway as major route for camptothecin biosynthesis in Nothapodytes nimmoniana (Graham) Mabb. BMC Plant Biol 19(1):1–15
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140
Rogowska A, Szakiel A (2020) The role of sterols in plant response to abiotic stress. Phytochem Rev 19(6):1525–1538
Sahakyan NZ, Ginovyan M, Petrosyan M et al (2019) Antibacterial and anti-phage activity of Plantago major L. raw material. Chem Biol 53(1):59–64
Samuelsen AB (2000) The traditional uses, chemical constituents and biological activities of Plantago major L. A review. J Ethnopharmacol 71(1–2):1–21
Schläpfer P, Zhang P, Wang C, Kim T et al (2017) Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol 173(4):2041–2059
Schluttenhofer C, Yuan L (2015) Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol 167(2):295–306
Shad AA, Ahmad S, Ullah R et al (2014) Phytochemical and biological activities of four wild medicinal plants. The Sci W J. https://doi.org/10.1155/2014/857363
Shahriari Z, Heidari B, Dadkhodaie A (2018) Dissection of genotype × environment interactions for mucilage and seed yield in Plantago species: application of AMMI and GGE biplot analyses. PLoS ONE 13(5):e0196095
Shirazi Z, Aalami A, Tohidfar M et al (2019) Triterpenoid gene expression and phytochemical content in Iranian licorice under salinity stress. Protoplasma 256(3):827–837
Suzuki H, Fukushima EO, Shimizu Y et al (2019) Lotus japonicus triterpenoid profile and characterization of the CYP716A51 and LjCYP93E1 genes involved in their biosynthesis in planta. Plant Cell Physiol 60(11):2496–2509
Thirumurugan D, Cholarajan A, Raja SS, Vijayakumar R (2018) An introductory chapter: secondary metabolites. Secondary metabolites: sources and applications 1–21. https://doi.org/10.5772/intecopen.79766
Upadhyay S, Jeena GS, Kumar (2020) Asparagus racemosus bZIP transcription factor-regulated squalene epoxidase (ArSQE) promotes germination and abiotic stress tolerance in transgenic tobacco. Plant Sci 290:110291
Wang DH, Du F, Liu HY et al (2010) Drought stress increases iridoid glycosides biosynthesis in the roots of Scrophularia ningpoensis seedlings. J Med Plants Res 4(24):2691–2699
Wei H, Xu C, Movahedi A et al (2019) Characterization and function of 3-Hydroxy-3-Methylglutaryl-CoA reductase in Populus trichocarpa: overexpression of PtHMGR enhances terpenoids in transgenic Poplar. Front Plant Sci 10:1476. https://doi.org/10.3389/fpls.2019.01476
Xu C, Jiao C, Zheng Y, Sun H, Liu W et al (2015) De novo and comparative transcriptome analysis of cultivated and wild spinach. Sci Rep 5:17706
Yadav RK, Sangwan RS, Sabir F (2014) Effect of prolonged water stress on specialized secondary metabolites, peltate glandular trichomes, and pathway gene expression in Artemisia annua L. Plant Physiol Biochem 74:70–83
Yadav RK, Sangwan RS, Srivastava AK (2017) Prolonged exposure to salt stress affects specialized metabolites-artemisinin and essential oil accumulation in Artemisia annua L.: metabolic acclimation in preferential favour of enhanced terpenoid accumulation accompanying vegetative to reproductive pha. Protoplasma 254:505–522
Yamamura Y, Kurosaki F, Lee JB (2017) Elucidation of terpenoid metabolism in Scoparia dulcis by RNA-seq analysis. Sci Rep 7(1):1–14
You MK, Lee YJ, Kim JK et al (2020) The organ-specific differential roles of rice DXS and DXR, the first two enzymes of the MEP pathway, in carotenoid metabolism in Oryza sativa leaves and seeds. BMC Plant Biol 20:1–16
Yuan Y, Zhang B, Tang X et al (2020) Comparative transcriptome analysis of different Dendrobium species reveals active ingredients-related genes and pathways. Int J Mol Sci 21(3):861
Zhang H, Mittal N, Leamy LJ, Barazani O, Song BH (2017) Back into the wild—apply untapped genetic diversity of wild relatives for crop improvement. Evol Appl 10:5–24
Zhang Z, Liu W, Ma Z et al (2019) Transcriptional characterization and response to defense elicitors of mevalonate pathway genes in cotton (Gossypium arboreum L). Peer J 7:e8123
Zhou C, Mei X, Rothenberg DON et al (2020a) Metabolome and transcriptome analysis reveals putative genes involved in anthocyanin accumulation and coloration in white and pink tea (Camellia sinensis) flower. Molecules 25(1):190
Zhou HC, Shamala LF, Yi XK (2020b) Analysis of terpene synthase family genes in Camellia sinensis with an emphasis on abiotic stress conditions. Sci Rep 10(1):1–13
Acknowledgements
SG acknowledge “National Postdoctoral Fellowship” scheme of the Science & Engineering Research Board (SERB) for their financial support. The authors would like to acknowledge the Director CSIR-Indian Institute of Integrative Medicine for providing the research facilities and infrastructure. We thank Negenome Biosolutions Pvt. Ltd. for their help. The authors are thankful to Amit Kumar, CSIR IIIM, Jammu for facilitating HPLC analyses.
Funding
The work is supported by DST SERB National Postdoctoral Fellowship (NPDF) award number PDF/2017/001741.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Communicated by Handling Editor: Peter Nick.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
709_2021_1663_MOESM1_ESM.docx
Supplementary file1: Sequence of primers used to amplify the genes of terpenoid and iridoid pathways using quantitative real-time PCR (qRT-PCR). (DOCX 13 KB)
709_2021_1663_MOESM2_ESM.xlsx
Supplementary file 2: Annotation of assembled transcripts satisfying the criteria of E-value (10-5) and minimum query coverage (40%) against Uniprot plant database (XLSX 11075 KB)
709_2021_1663_MOESM3_ESM.xlsx
Supplementary file 3: BLASTX hit of unigenes of P.major and P. ovata showing sequence homology with NCBI genomes (XLSX 20 KB)
709_2021_1663_MOESM4_ESM.xlsx
Supplementary file 4: Gene ontology annotation information of differentially expressed unigenes (biological process; sheet 1, cellular components; sheet 2, molecular functions; sheet 3) (XLSX 82 KB)
709_2021_1663_MOESM5_ESM.xlsx
Supplementary file 5: KEGG pathway analysis of 526 differentially expressed unigenes. Pathways are arranged in descending order of unigenes number. (XLSX 10 KB)
709_2021_1663_MOESM6_ESM.xlsx
Supplementary file 6: List of upregulated genes in P.major in comparison to P. ovata with their fold change value and general gene information retrieved from Uniprot database. (XLSX 641 KB)
709_2021_1663_MOESM7_ESM.xlsx
Supplementary file 7: List of downregulated genes in P.major in comparison to P. ovata with their fold change value and general gene information retrieved from Uniprot database. (XLSX 605 KB)
709_2021_1663_MOESM8_ESM.jpg
Supplementary Fig. 1: Heat map showing comparative expression analysis of genes related to terpenoid (20 genes) and iridoid (10 genes) pathways in two species of Plantago (P. ovata and P. major) based on FPKM values. Colors correspond with expression values; Dark pink indicates low expression, red indicates average expression, and light pink color indicates high expression levels. (JPG 44 KB)
Rights and permissions
About this article
Cite this article
Gupta, S., Singh, R., Sharma, A. et al. Comparative transcriptome mining for terpenoid biosynthetic pathway genes in wild and cultivated species of Plantago. Protoplasma 259, 439–452 (2022). https://doi.org/10.1007/s00709-021-01663-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-021-01663-9