Abstract
Mastigonemes, tripartite tubular hairs on the anterior flagellum of Phytophthora zoospores, are instrumental for disease dissemination to new host plants. A previous study showed that PnMas2 was part of the tubular shaft of Phytophthora parasitica mastigonemes. In the current study, genes encoding two related proteins, PnMas1 and PnMas3, were identified in the genome of P. parasitica. PnMas1 interacts with PnMas2 and also occurs along the mastigoneme shaft. RNA-Seq analyses indicate that PnMas1 and PnMas2 genes have similar expression profiles both in vitro and in planta but that PnMas3 is expressed temporally prior to PnMas1 and PnMas2 during asexual development and plant infection. Immunocytochemistry and GFP-tagging document the occurrence of all three PnMas proteins within the specialised compartments of the ER during mastigoneme formation, but only PnMas1 and PnMas2 occur in mature mastigonemes on the flagellar surface. Anti-PnMas1 and anti-PnMas2 antibodies co-labelled two high-molecular-weight (~400 kDa) protein complexes in native gels but anti-PnMas3 antibodies labelled a 65 kDa protein complex. Liquid chromatography-mass spectrometry analysis identified PnMas1 and PnMas2 but not PnMas3 in flagellar extracts. These results suggest that PnMas3 associates with mastigonemes during their assembly within the ER but is not part of mature mastigonemes on the anterior flagellum. Phylogenetic analyses using homologues of Mas genes from the genomes of 28 species of Stramenopiles give evidence of three Mas sub-families, namely Mas1, Mas2 and Mas3. BLAST analyses showed that Mas genes only occur in flagellate species within the Stramenopile taxon.







Similar content being viewed by others
References
Ah-Fong AM, Judelson HS (2011) Vectors for fluorescent protein tagging in Phytophthora: tools for functional genomics and cell biology. Fungal Biol 115:882–890. https://doi.org/10.1016/j.funbio.2011.07.001
Aoki T, Tahara T, Satoh K, Fujino H, Watabe H (2003) General properties of GFP-display, an electrophoretic analysis for single amino acid changes in target polypeptides. Anal Biochem 317:107–115. https://doi.org/10.1016/S0003-2697(03)00112-X
Aoki T, Takahashi Y, Koch KS, Leffert HL, Watabe H (1996) Construction of a fusion protein between protein a and green fluorescent protein and its application to Western blotting. FEBS Lett 384:193–197. https://doi.org/10.1016/0014-5793(96)00289-X
Armbrust EV (1999) Identification of a new gene family expressed during the onset of sexual reproduction in the centric diatom Thalassiosira weissflogii. Appl Environ Microbiol 65:3121–3128
Armbrust EV, Galindo HM (2001) Rapid evolution of a sexual reproduction gene in centric diatoms of the genus Thalassiosira. Appl Environ Microbiol 67:3501–3513. https://doi.org/10.1128/AEM.67.8.3501-3513.2001
Australian Government Department of the Environment (2014) Threat abatement plan for disease in natural ecosystem caused by Phyotophthora cinnamomi. Australian Government Department of the Environment. http://www.environment.gov.au/resource/threat-abatement-plan-disease-natural-ecosystems-caused-phytophthora-cinnamomi Accessed March 2015
Balci, Y, Bienapfl, JC & Lamour, K (2013) Phytophthora in US forests. In: Lamour, K (ed.) Phytophthora: a global perspective. CABI, Croydon, pp 135–145
Blackman LM, Arikawa M, Yamada S, Suzaki T, Hardham AR (2011) Identification of a mastigoneme protein from Phytophthora nicotianae. Protist 162:100–114. https://doi.org/10.1016/j.protis.2010.01.005
Blackman LM, Cullerne DP, Torreña P, Taylor J, Hardham AR (2015) RNA-Seq analysis of the expression of genes encoding cell wall degrading enzymes during infection of lupin (Lupinus angustifolius) by Phytophthora parasitica. PLoS One 10:e0136899. https://doi.org/10.1371/journal.pone.0136899
Bottin A, Larche L, Villalba F, Gaulin E, Esquerré-Tugayé M-T, Rickauer M (1999) Green fluorescent protein (GFP) as gene expression reporter and vital marker for studying development and microbe-plant interaction in the tobacco pathogen Phytophthora parasitica var. nicotianae. FEMS Microbiol Lett 176:51–56. https://doi.org/10.1016/S0378-1097(99)00218-9
Bouck GB (1969) Extracellular microtubules. The origin, structure, and attachment of flagellar hairs in Fucus and Ascophyllum antherozoids. J Cell Biol 40:446–460
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brasier C, Denman S, Brown A, Webber J (2004) Sudden oak death (Phytophthora ramorum) discovered on trees in Europe. Mycol Res 108:1108–1110
Cahill DM, Cope M, Hardham AR (1996) Thrust reversal by tubular mastigonemes: immunological evidence for a role of mastigonemes in forward motion of zoospores of Phytophthora cinnamomi. Protoplasma 194:18–28. https://doi.org/10.1007/Bf01273164
Cameron JN, Carlile MJ (1978) Fatty acids, aldehydes and alcohols as attractants for zoospores of Phytophthora palmivora. Nature 271:448–449. https://doi.org/10.1038/271448a0
Chen LL, Haines TH (1976) The flagellar membrane of Ochromonas danica. Isolation and electrophoretic analysis of the flagellar membrane, axonemes, and mastigonemes. J Biol Chem 251:1828–1834
Cope M, Hardham A (1994) Synthesis and assembly of flagellar surface antigens during zoosporogenesis in Phytophthora cinnamomi. Protoplasma 180:158–168. https://doi.org/10.1007/Bf01507852
D'Amico F, Skarmoutsou E, Stivala F (2009) State of the art in antigen retrieval for immunohistochemistry. J Immunol Methods 341:1–18. https://doi.org/10.1016/j.jim.2008.11.007
Erwin DC & Ribeiro OK (1996) Phytophthora disease worldwide. The American Phytopathological Society Press, St. Paul, Minnesota, USA
Fry WE, Goodwin SB (1997) Resurgence of the Irish potato famine fungus. Bioscience 47:363–371. https://doi.org/10.2307/1313151
Fu G, Nagasato C, Oka S, Cock JM, Motomura T (2014) Proteomics analysis of heterogeneous flagella in brown algae (Stramenopiles). Protist 165:662–675. https://doi.org/10.1016/j.protis.2014.07.007
Gubler F, Hardham A, Duniec J (1989) Characterising adhesiveness of Phytophthora cinnamomi zoospores during encystment. Protoplasma 149:24–30. https://doi.org/10.1007/Bf01623979
Hardham A, Gubler F, Duniec J, Elliott J (1991) A review of methods for the production and use of monoclonal antibodies to study zoosporic plant pathogens. J Microsc 162:305–318. https://doi.org/10.1111/j.1365-2818.1991.tb03142.x
Hardham AR (1985) Studies on the cell surface of zoospores and cysts of the fungus Phytophthora cinnamomi: the influence of fixation on patterns of lectin binding. J Histochem Cytochem 33:110–118. https://doi.org/10.1177/33.2.3918095
Hardham AR (1987) Microtubules and the flagellar apparatus in zoospores and cysts of the fungus Phytophthora cinnamomi. Protoplasma 137:109–124. https://doi.org/10.1007/BF01281146
Hardham AR, Blackman LM (2018) Phytophthora cinnamomi. Mol Plant Pathol 19:260–285. https://doi.org/10.1111/mpp.12568
Hardham AR, Gubler F (1990) Polarity of attachment of zoospores of a root pathogen and pre-alignment of the emerging germ tube. Cell Biol Int Rep 14:947–956. https://doi.org/10.1016/0309-1651(90)90107-a
Hardham AR, Hyde GJ (1997) Asexual sporulation in the oomycetes. Adv Bot Res 24:353–398
Haseloff J (1999) GFP variants for multispectral imaging of living cells. Methods Cell Biol 58:139–151
Heath IB, Greenwood AD, Griffiths HB (1970) The origin of Flimmer in Saprolegnia, Dictyuchus, Synura and Cryptomonas. J Cell Sci 7:445–461
Hee W, Torreña P, Blackman L, Hardham A & Lamour K (2013) Phytophthora cinnamomi in Australia. In: Lamour, K (ed.) Phytophthora: a global perspective. CABI, Croydon, pp 124–134
Hill FG, Outka DE (1974) The structure and origin of mastigonemes in Ochromonas minute and Monas sp. J Protozool 21:299–312
Hogg PJ (2003) Disulfide bonds as switches for protein function. Trends Biochem Sci 28:210–214. https://doi.org/10.1016/S0968-0004(03)00057-4
Honda D, Shono T, Kimura K, Fujita S, Iseki M, Makino Y, Murakami A (2007) Homologs of the sexually induced gene 1 (sig1) product constitute the stramenopile mastigonemes. Protist 158:77–88. https://doi.org/10.1016/j.protis.2006.08.004
Jahn, TL, Lanoman, MD & Fonseca, JR (1964) The mechanism of locomotion of flagellates. II. Function of the mastigonemes of Ochromonas*. 11, 291–296
Judelson HS, Shrivastava J, Manson J (2012) Decay of genes encoding the oomycete flagellar proteome in the downy mildew Hyaloperonospora arabidopsidis. PLoS One 7:e47624. https://doi.org/10.1371/journal.pone.0047624
Judelson HS, Tyler BM, Michelmore RW (1991) Transformation of the oomycete pathogen, Phytophthora infestans. Mol Plant-Microbe Interact 4:602–607
Kamoun S, Furzer O, Jones JD, Judelson HS, Ali GS, Dalio RJ, Roy SG, Schena L, Zambounis A, Panabieres F, Cahill D, Ruocco M, Figueiredo A, Chen XR, Hulvey J, Stam R, Lamour K, Gijzen M, Tyler BM, Grunwald NJ, Mukhtar MS, Tome DF, Tor M, Van Den Ackerveken G, McDowell J, Daayf F, Fry WE, Lindqvist-Kreuze H, Meijer HJ, Petre B, Ristaino J, Yoshida K, Birch PR, Govers F (2015) The top 10 oomycete pathogens in molecular plant pathology. Mol Plant Pathol 16:413–434. https://doi.org/10.1111/mpp.12190
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Latijnhouwers M, Ligterink W, Vleeshouwers V, van West P, Govers F (2004) A G alpha subunit controls zoospore motility and virulence in the potato late blight pathogen Phytophthora infestans. Mol Microbiol 51:925–936. https://doi.org/10.1046/j.1365-2958.2003.03893.x
Ludowici VA, Zhang W, Blackman LM, Hardham AR, Lamour K (2013) Phytophthora nicotianae. In: Lamour K (ed) Phytophthora: a global perspective. CABI, Croydon, pp 113–123
Markey DR, Bouck GB (1977) Mastigoneme attachment in Ochromonas. J Ultrastruct Res 59:173–177
Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F (2009) TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25:126–127. https://doi.org/10.1093/bioinformatics/btn575
Morris BM, Gow NAR (1993) Mechanism of electrotaxis of zoospores of phytopathogenic fungi. Phytopathology 83:877–882. https://doi.org/10.1094/Phyto-83-877
Morris BM, Reid B, Gow NAR (1992) Electrotaxis of zoospores of Phytophthora palmivora at physiologically relevant field strengths. Plant Cell Environ 15:645–653. https://doi.org/10.1111/j.1365-3040.1992.tb01006.x
Morris PF, Ward EWB (1992) Chemoattraction of zoospores of the soybean pathogen, Phytophthora sojae, by isoflavones. Physiol Mol Plant P 40:17–22. https://doi.org/10.1016/0885-5765(92)90067-6
Narayan RD, Blackman LM, Shan W, Hardham AR (2010) Phytophthora nicotianae transformants lacking dynein light chain 1 produce non-flagellate zoospores. Fungal Genet Biol 47:663–671. https://doi.org/10.1016/j.fgb.2010.04.008
Perkins DN, Pappin DJC, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. https://doi.org/10.1002/(sici)1522-2683(19991201)20:18<3551::aid-elps3551>3.0.co;2-2
Rizzo DM, Garbelotto M (2003) Sudden oak death: endangering California and Oregon forest ecosystems. Front Ecol Environ 1:197–204. https://doi.org/10.2307/3868064
Robideau GP, Rodrigue N, André Lévesque C (2014) Codon-based phylogenetics introduces novel flagellar gene markers to oomycete systematics. Mol Phylogenet Evol 79:279–291
Robold A, Hardham A (1998) Production of species-specific monoclonal antibodies that react with surface components on zoospores and cysts of Phytophthora nicotianae. Can J Microbiol 44:1161–1170. https://doi.org/10.1139/cjm-44-12-1161
Robold AV, Hardham AR (2005) During attachment Phytophthora spores secrete proteins containing thrombospondin type 1 repeats. Curr Genet 47:307–315
Shan WX, Marshall JS, Hardham AR (2004) Gene expression in germinated cysts of Phytophthora nicotianae. Mol Plant Pathol 5:317–330. https://doi.org/10.1111/j.1364-3703.2004.00231.x
van West P, Morris BM, Reid B, Appiah AA, Osborne MC, Campbell TA, Shepherd SJ, Gow NAR (2002) Oomycete plant pathogens use electric fields to target roots. Mol Plant-Microbe Interact 15:790–798. https://doi.org/10.1094/mpmi.2002.15.8.790
Yamagishi T, Motomura T, Nagasato C, Kato A, Kawai H (2007) A tubular mastigoneme-related protein, Ocm1, isolated from the flagellum of a chromophyte alga, Ochromonas danica. J Phycol 43:519–527. https://doi.org/10.1111/j.1529-8817.2007.00340.x
Yamagishi T, Motomura T, Nagasato C, Kawai H (2009) Novel proteins comprising the Stramenopile tripartite mastigoneme in Ochromonas danica (Chrysophyceae). J Phycol 45:1110–1115. https://doi.org/10.1111/j.1529-8817.2009.00722.x
Yang X, Tyler BM, Hong C (2017) An expanded phylogeny for the genus Phytophthora. IMA fungus 8:355–398. https://doi.org/10.5598/imafungus.2017.08.02.09
Zadoks J (2008) The potato murrain on the European continent and the revolutions of 1848. Potato Res 51:5–45. https://doi.org/10.1007/s11540-008-9091-4
Zhang W, Blackman LM, Hardham AR (2013) Transient fusion and selective secretion of vesicle proteins in Phytophthora nicotianae zoospores. PeerJ 1:e221. https://doi.org/10.7717/peerj.221
Acknowledgements
The authors would like to thank Dr. Thy Truong (ANU) for assistance with the LC-MS/MS and Professor Richard W. Michelmore (UC Davis) for providing analyses of unpublished sequence data from Bremia lactucae. We would also like to thank Simon Michnowicz and Eugene Kapp from Walter and Eliza Hall Institute of Medical Research for incorporating genomic data of P. parasitica into the Mascot database. We also thank Dr. Megan Mcdonald for advice on phylogenetics analysis. The work was supported by grants from The Hermon Slade Foundation and the Australian Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Ulrike Mathesius
Electronic supplementary material
Online Resource 1
Primer sequences used to construct the PnMas3-GFP transgene. (DOCX 12 kb)
Online Resource 2
Light micrographs of samples collected during P. parasitica flagella isolation. (a) Immunofluorescence labelling with anti-PnMas2 mAb shows that mastigonemes are still attached to the anterior flagellum (arrowheads) after the flagella isolation. (b) The posterior flagella which are not labelled by anti-PnMas2 mAb are visible using DIC optics (arrows). Scale bars = 20 μm. (PNG 476 kb)
Online Resource 3
Alignment of PnMas1, PnMas2 and PnMas3 protein sequences. All three Mas proteins contain conserved cysteine residues (cyan) within EGF-like domains (yellow). (PNG 1000 kb)
Online Resource 4
Statistical analysis of PnMas1, PnMas2 and PnMas3 transcript levels at six stages in the P. parasitica asexual life cycle. (DOCX 13 kb)
Online Resource 5
Details of the generation of monoclonal antibodies (mAbs) against P. parasitica PnMas1 and PnMas3 proteins. The table shows the mAb cell lines, epitopes targeted by the mAbs and the results of immunoblotting and immunocytochemical labelling. (DOCX 13 kb)
Online Resource 6
The results of immunofluorescence labelling of P. parasitica zoospores with anti-PnMas1 and anti-PnMas3 mAbs after different fixation regimes. (DOCX 12 kb)
Online Resource 7
Micrographs of P. parasitica zoospores fixed in 80% methanol and labelled with anti-PnMas1 mAbs and GAM-FITC. a-e: mAbs 2 K19, 2 K23, 2 M19, 2A21 and 2 L5 label mastigonemes on the anterior flagellum and in ER packets within the cells. f: mAb 2H18 did not show any labelling of the zoospores. Scare bars = 5 μm (PNG 1184 kb)
Online Resource 8
Brightfield (a, c, e, g, i) and fluorescence (b, d, f, h, j) micrographs showing P. parasitica zoospores labelled with anti-PnMas1 mAb (2 M19) and GAM-FITC. The zoospores were treated with 80% methanol (a, b), 80% acetone (c, d), 63% ethanol/3% acetic acid (e, f), 0.05% Triton X-100 (g, h) and 0.05% Tween 20 (i, j). The anti-PnMas1 mAb labels mastigonemes on the anterior flagella (arrows) but does not label the posterior flagella (arrowheads). Methanol treated zoospores often round up and the flagella adhered to the cell surface (b). Scale bars = 5 μm. (PNG 1102 kb)
Online Resource 9
Immunoblots of P. parasitica proteins separated using native PAGE with anti-PnMas1 (2 M19), anti-PnMas2 and anti-PnMas3 (2O20) mAbs. Both anti-PnMas1 and anti-PnMas2 mAbs react with a pair of bands ~400 kDa in size (arrows). Anti-PnMas3 mAb cell line 2O20 labelled a 65 kDa protein complex (arrowhead). (PNG 148 kb)
Online Resource 10
LC-MS/MS analysis of proteins co-precipitated by anti-PnMas2 mAb in sporulating hyphae extracts. (DOCX 14 kb)
Online Resource 11
LC-MS/MS analysis of proteins co-precipitated by anti-PnMas2 mAb in P. parasitica flagellar protein extracts. (DOCX 16 kb)
Online Resource 12
Accession number of Mas genes. (DOCX 16 kb)
Online Resource 13
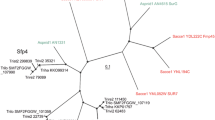
MrBayes analysis of the phylogenetic relationships of concatenated Mas1, Mas2 and Mas3 protein sequences from putative homologues from the Stramenopile genomes. The EGF-like domains and N-termini of the proteins were retained for the analysis. The numbers on the branches show the posterior probabilities generated from 10,000 generations. The locations of PnMas proteins are indicated by the arrows. (PNG 748 kb)
Online Resource 14
A phylogenetic tree showing neighbour joining analysis of six heat shock protein 70s from Chlamydomonas reinhardtii (Crhsp70A–F) and 12 hsp70-like proteins identified in the P. parasitica genome. The branches show bootstrap values generated from 10,000 replicates. The analysis showed that PPTG_02122 (blue arrow), which was detected in the LC-MS/MS analysis of anti-PnMas2 co-precipitated proteins and in P. parasitica isolated flagellar extracts, is closely related to Crhsp70A (red arrow), a protein found in Chlamydomonas flagella. (DOCX 1539 kb)
Rights and permissions
About this article
Cite this article
Hee, W.Y., Blackman, L.M. & Hardham, A.R. Characterisation of Stramenopile-specific mastigoneme proteins in Phytophthora parasitica. Protoplasma 256, 521–535 (2019). https://doi.org/10.1007/s00709-018-1314-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-018-1314-1