Abstract
Plant sap-feeding hemipterans harbor obligate symbiotic microorganisms which are responsible for the synthesis of amino acids missing in their diet. In this study, we characterized the obligate symbionts hosted in the body of the xylem-feeding leafhopper Evacanthus interruptus (Cicadellidae: Evacanthinae: Evacanthini) by means of histological, ultrastructural and molecular methods. We observed that E. interruptus is associated with two types of symbiotic microorganisms: bacterium ‘Candidatus Sulcia muelleri’ (Bacteroidetes) and betaproteobacterium that is closely related to symbionts which reside in two other Cicadellidae representatives: Pagaronia tredecimpunctata (Evacanthinae: Pagaronini) and Hylaius oregonensis (Bathysmatophorinae: Bathysmatophorini). Both symbionts are harbored in their own bacteriocytes which are localized between the body wall and ovaries. In E. interruptus, both Sulcia and betaproteobacterial symbionts are transovarially transmitted from one generation to the next. In the mature female, symbionts leave the bacteriocytes and gather around the posterior pole of the terminal oocytes. Then, they gradually pass through the cytoplasm of follicular cells surrounding the posterior pole of the oocyte and enter the space between them and the oocyte. The bacteria accumulate in the deep depression of the oolemma and form a characteristic ‘symbiont ball’. In the light of the results obtained, the phylogenetic relationships within modern Cicadomorpha and some Cicadellidae subfamilies are discussed.
Similar content being viewed by others
Introduction
The Hemiptera are a large group of insects with feeding habits that range from phytophagy to predation, including ectoparasitism and hematophagy (Forero 2008). Plant feeders suck the phloem or xylem sap, or ingest plant cell content (Backus 1988; Campbell et al. 1994; Sorensen et al. 1995). Among the six suborders of the Hemiptera (i.e. Paleorrhyncha,Sternorrhyncha, Fulgoromorpha, Cicadomorpha, Coleorrhyncha and Heteroptera; Szwedo et al. 2004; Grimaldi and Engel 2005; Drohojowska et al. 2013), the Cicadomorpha: Clypeata lineage (uniting extinct Hylicelloidea and extant Cicadoidea, Cercopoidea and Membracoidea sensu lato) is the only one with several strong adaptations for xylem feeding (Wang et al. 2012), however in some of them shifts to cell-content and back to phloem feeding occurred (Dietrich 2013). The diet of xylem-feeding hemipterans is extremely unbalanced, because the amount of nutrients in the xylem sap is 10 times less than that in the phloem (Andersen et al. 1989). Therefore, most plant sup-sucking hemipterans live in mutualistic associations with symbiotic microorganisms (bacteria or yeast) which synthesize missing nutrients and provide them to their hosts (Wilkinson and Ishikawa 2001; Baumann 2005; Douglas 2009). The sequencing of the genome of bacteria inhabiting the body of hemipterans has indicated that these microorganisms possess the biosynthetic pathways necessary for the synthesis of essential amino acids, i.e. leucine, serine, tryptophan as well as many vitamins and other cofactors (Douglas 2006; Wu et al. 2006; McCutcheon and Moran 2007; McCutcheon et al. 2009).
The obligate symbionts may be localized both extracellularly, in the lumen of midgut appendages (in some heteropterans), and intracellularly, in cells of the midgut epithelium (in some heteropterans) or in specialized cells of mesodermal origin termed bacteriocytes/mycetocytes (in most hemipterans) (see Kikuchi 2009 for further details). The latter microorganisms are termed ‘mycetomic symbionts’.
Among hemipterans, the Sternorrhyncha (aphids, psyllids, whiteflies, scale insects) are associated with one type of obligate mycetomic symbiont (termed ‘primary symbiont’), e.g. aphids harbor the bacterium Buchnera aphidicola, psyllids – the bacterium Carsonella ruddii (see Baumann 2005 for further details). Apart from obligate symbionts, sternorrhynchans, as a rule, hold additional symbionts (termed secondary or facultative symbionts) that may play various roles for their host insects, e.g. may protect them from infection by fungal pathogens or attack by parasitic hymenopterans (Oliver et al. 2003; Scarborough et al. 2005). In contrast to the situation observed in Sternorrhyncha, in Cicadomorpha and Fulgoromorpha (both formerly treated as Auchenorrhyncha), two or more types of obligate symbionts (termed ‘coprimary symbionts’) co-occur and all of them are engaged in the synthesis of nutrients essential to the host insect (Moran et al. 2003; Takiya et al. 2006; Bressan et al. 2009; Noda et al. 2012; Urban and Cryan 2012; Bennett and Moran 2013; Ishii et al. 2013; Koga et al. 2013; Michalik et al. 2014a; Wu et al. 2006).
Molecular analyses have shown that both Cicadomorpha and Fulgoromorpha usually possess the obligate Bacteroidetes bacterium ‘Candidatus Sulcia muelleri’ (hereafter referred to as Sulcia) and one other type of coprimary symbiont, e.g. gammaproteobacterium ‘Candidatus Baumannia cicadellinicola’ (hereafter Baumannia), betaproteobacterium ‘Candidatus Zinderia insecticola’ (hereafter Zinderia), betaproteobacterium ‘Candidatus Vidania fulgoroideae’ (hereafter Vidania), betaproteobacterium ‘Candidatus Nasuia deltocephalinicola’ (hereafter Nasuia), alphaproteobacterium ‘Candidatus Hodgkinia cicadicola’ (hereafter Hodgkinia) (Moran et al. 2003, 2005; Takiya et al. 2006; McCutcheon et al. 2009; Gonella et al. 2011; Noda et al. 2012; Urban and Cryan 2012; Ishii et al. 2013; Koga et al. 2013). The results of recent molecular phylogenetic analyses (Bennett and Moran 2013; Koga et al. 2013; Bennett et al. 2014; Koga and Moran 2014) suggest that this enormous diversity of symbionts found in Cicadomorpha and Fulgoromorpha results from a multiple, independent replacement of symbiotic bacteria by other microorganisms. An ancestor of Cicadomorpha and Fulgoromorpha has been colonized by Sulcia and betaproteobacterial symbionts (probably 270 million years ago), but during the further evolution of the hemipteran lineages the betaproteobacterial co-symbiont was replaced by other bacteria (Moran et al. 2005; Bennett and Moran 2013; Koga et al. 2013; Koga and Moran 2014). In some planthopper families, i.e. Flatidae and Delphacidae as well as in leafhopper Scaphoideus titanus (Cicadellidae: Deltocephalinae) bacterial symbionts have been replaced by yeast symbionts (Sacchi et al. 2008; Michalik et al. 2009; Noda 1977).
On account of their crucial role, the obligate symbionts of hemipterans are transovarially (vertically) transmitted between generations (see Buchner 1965 for further details). For this reason, bacteria living in different species of insects evolved independently from one another, i.e. without gene exchange between bacteria in different hosts.
Evacanthus interruptus (Linnaeus, 1758) is a representative of leafhoppers (Cicadomorpha: Membracoidea: Cicadellidae). So far, the symbiotic microorganisms present in the xylem-feeding subfamily Evacanthinae Metcalf, 1939 have been examined only fragmentarily (Buchner 1965; Takiya et al. 2006). Early microscopic observations conducted by Buchner (1965) showed that the females of Evacanthus interruptus possess two types of symbiotic microorganisms, which are localized in separate bacteriocytes. More recently, using molecular methods, Takiya et al. (2006) indicated the occurrence of Sulcia symbiont in the Pagaronia tredecimpunctata species Ball, 1902. Koga et al. (2013) detected betaproteobacterial symbionts in the same leafhopper species.
The objectives of this study were to examine the ultrastructure of symbionts of E. interruptus, their distribution in the host insect body, mode of transovarial transmission from one generation to the next and to determine their systematic affinity.
Material and methods
Insects
Adult females of Evacanthus interruptus (Linnaeus, 1758) were collected from herbaceous plants in Kraków (Poland) and near Vorokhta (Chornohora Mountain, Eastern Carpathians, Ukraine), from July to September.
DNA analyses
The dissected bacteriomes were fixed in 96 % ethanol, washed twice in sterile water and homogenized in 120 μl of 0.7 M NH4OH. After 15 min of incubation at 100 °C in alkaline conditions, tubes were opened and further incubated at 100 °C for 10 min. Next, the samples were centrifuged (5 min, 12 000 rpm) and supernatants were transferred to the new tubes. Two volumes of 96 % ethanol and 1/10 volume of 3 M sodium acetate were added to every tube. DNA precipitations were carried out for 30 min at −20 °C. After centrifugation (15 min, 12 000 rpm, 4 °C), DNA pellets were washed in 250 μl of 70 % ethanol and centrifuged (15 min, 12 000 rpm, room temperature). The DNA was then dried for 15 min at 37 °C, dissolved in 20 μl of sterile water, and stored at −20 °C for further analysis.
The polymerase chain reaction (PCR) for the detection of the symbionts harbored in the bacteriocytes of E. interruptus were performed using the following specific primers: 10CFBFF (5’AGAGTTTGATCATGGCTCAGGATG3’) and 1515R (5’GTACGGCTACCTTGTTACGACTTAG3’) for Sulcia symbiont (Moran et al. 2005); 16SA1 (5’AGAGTTTGATCMTGGCTCAG3’) and Bet940R (5’TTAATCCACATCATCCACCG3’) (Koga et al. 2013) for betaproteobacterial symbiont, as well as 10 F (5’AGTTTGATCATGGCTCAGATTG3’) and 650R (5’CACCGGTACATATGAAATTCT3’) for Baumannia symbiont (Takiya et al. 2006). The PCR mixture contained 2 μl of 10x PCR buffer, 0.3 μl of 25 mM MgCl2, 0.5 μl of 10 mM deoxynucleotide triphosphate (dNTP) mixture, 0.5 μl of each 10 mM forward and reverse primers, 2 μl of 1000 ng/μl extracted DNA, 1.25U of JumpStart™ Taq DNA polymerase (Sigma-Aldrich, Germany) and 12.7 μl of sterile, deionized water. The PCR profile consisted of an initial denaturation step at 94 °C for 3 min, followed by 33 cycles at 94 °C for 30 s, 50 °C (for primers:10 F and 650R), 52 °C (for primers: 16SA1 and Bet940R) or 55 °C (for primers:10CFBFF and 1515R) for 40 s, 70 °C for 1 min 40 s and a final extension step of 5 min at 72 °C. PCR products were subjected to electrophoresis on 1 % agarose gel stained with ethidium bromide and purified using the GenElute MinusEtBr Spin Columns (Sigma-Aldrich, Germany). The purified amplification products were used for sequencing in two directions. The obtained sequences were deposited in GenBank database with accession numbers KP278014–KP278015 and KP943506–KP943507.
Phylogenetic analyses
The sequences of the 16S rDNA of symbiotic microorganisms used in comprehensive phylogenetic analyses were downloaded from GenBank. The sequences were edited using BioEdit Sequence Alignment Editor 5.0.9 (Hall 1999), and aligned using ClustalX 1.8 (Thompson et al. 1997). All alignments were verified and corrected visually. For the analyses, the appropriate nucleotide substitution model was first determined using Modeltest 3.06 (Posada and Crandall 1998). Based on fossil evidence (Moran et al. 2005; Bennett and Moran 2013; Koga et al. 2013; Koga and Moran 2014) that calibration points required for the molecular dating analyses were assigned as follows: the Cicadomorpha crown clade was fixed at 270 MYA. Molecular clock analyses were conducted using 16S rDNA. Beast v 1.6.1 (Rambaut and Drummond 2007) was used to infer divergence times of lineages under a Bayesian statistical framework. The time of lineage divergence was estimated using the uncorrelated lognormal relaxed clock model. For each matrix, Beast was run using a Yule speciation process and two independent MCMC chains for 4 000 000 generations, sampling every 100th generation. The convergence to stationary and the effective sample size of the model parameters were checked using Tracer. The maximum clade-credibility trees were built with TreeAnnotator (Rambaut and Drummond 2007). FigTree 1.4.0 software (Rambaut 2008) was used to visualize the results including confidence intervals. Pairwise, genetic distances were computed using Mega 6.0 (Tamura et al. 2013).
Light and electron microscopy
Entire abdomens, dissected ovaries and bacteriomes destined for histological and ultrastructural studies were fixed in 2.5 % glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) at room temperature. The material was next rinsed in the phosphate buffer with an addition of 5.8 % sucrose and postfixed in 1 % osmium tetroxide (in the same buffer). After undergoing dehydration in a graded series of ethanol and acetone, the material was embedded in epoxy resin Epon 812 (Serva, Germany). Semithin sections (1 μm thick) were stained with 1 % methylene blue in 1 % borax and examined and photographed under light microscopes, Leica DMR and Nikon Eclipse 80i. Ultrathin sections (90 nm thick) were contrasted with lead citrate and uranyl acetate and analyzed and photographed in the Jeol JEM 100 SX and Jeol JEM 2100 (Jeol, Japan) electron microscopes.
Fluorescence microscopy
The dissected ovaries destined for histochemical studies were fixed in 4 % formaldehyde in phosphate buffered saline (PBS) for 45 min at room temperature, rinsed in the same buffer, dehydrated in a graded series of ethanol, embedded in Histocryl (Agar Scientific LTD, Stansted, UK) and cut into semithin sections (0.7 μm thick). The sections were then stained with DAPI (1 μg/ml; Sigma Chemical Co., St. Louis, USA) for 30 min in darkness at room temperature. Next, the material was examined and photographed under an Axiovert 200 M confocal microscope.
Results
Molecular identification and phylogenetic analysis of symbionts of Evacanthus interruptus
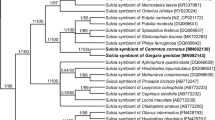
The systematic affinity of bacteriome-associated symbionts of E. interruptus from both Polish and Ukrainian populations was determined based on the analysis of their 16S rDNA sequences. In both samples (from Poland and Ukraine) the direct PCR revealed the presence of bacteria representing the phylum Bacteroidetes (bacterium Sulcia) and class Betaproteobacteria of the phylum Proteobacteria. The specific PCR has not revealed the occurrence of bacterium Baumannia in examined species (the specific product was not obtained). A comparison of the obtained 16S rDNA sequences of Sulcia symbionts from Polish and Ukrainian populations of E. interruptus indicated that they are identical, whereas there is a slight difference between the 16S rDNA sequences of betaproteobacterial symbionts (1 %). The length of the analyzed sequences was 960 pb for Sulcia and 678 pb for the betaproteobacterial symbiont, respectively. The mean base compositions of Sulcia symbionts were as follows: 30.8 % A, 25.2 % T, 17.3 % C, 26.6 % G, while in the betaproteobacterial symbionts there were: 33.2 % A, 25.9 % T, 15.7 % C, 25.1 % G. The variation within the 16S rDNA was about 12 % variable sites and 9 % parsimony informative sites for Sulcia symbionts and about 49 % variable sites and 43 % parsimony informative sites for the betaproteobacteria ones. BLASTN searches against the bacterial 16S rDNA sequences deposited in the GenBank clearly demonstrate the similarity of Bacteroidetes symbiont to the bacterium Sulcia isolated from the leafhopper, Pagaronia tredecimpunctata (99 %, 1217/1227) [AY676911], whereas the betaproteobacterial symbiont displays the highest similarity to the symbiont found in the two leafhoppers: Pagaronia tredecimpunctata (Evacanthinae: Pagaronini Anufriev, 1978) (89 %, 430/480) [AB772227] and Hylaius oregonensis (Baker, 1898) (Bathysmatophorinae Anufriev, 1978: Bathysmatophorini Anufriev, 1978) (90 %, 439/491) [AB772226]. The topology resulting from the Bayesian inference of the Sulcia and betaproteobacterial symbiont are shown in Figs. 1 and 2, respectively. Phylogenetic analyses of 16S rDNA of Sulcia symbionts of various representatives of Cicadomorpha (Fig. 1) show the monophyly of symbionts of the Membracoidea within Cicadomorpha: Clypeata (with 0.88 posterior probability values). Phylogenetic analyses of the 16S rDNA of the betaproteobacterial symbionts indicate that the symbionts of Evacanthinae (Pagaronia and Evacanthus) and Hylaius oregonensis (Bathysmatophorinae) form a monophyletic group which is closely related to betaproteobacterial symbionts of members of the Deltocephalinae leafhoppers (Fig. 2).
The phylogeny of the Sulcia symbionts of the extant Cicadomorpha: Clypeata based on a Bayesian analysis of partial 16S rDNA sequences. Dated tree generated using an uncorrelated longnormal clock in Beast. Posterior probabilities are indicated above the branches, whereas the numbers below the branches show the time of lineages divergence, scale is given with absolute geological ages (million years ago, MYA) (GenBank accession numbers of 16S rRNA genes are given in brackets). (Abbreviations for generic names: A. quadrinotata – Aphrophora quadrinotata; C. costalis – Cuerna costalis; C. major – Cyrtodisca major; C. striata – Cuerna striata; C. orni – Cicada orni; C. vicina – Chaetophyes vicina; C. viridis – Cicadella viridis; D. excisum – Diestostemma excisum; D. maidis – Dalbulus maidis; E. interruptus – Evacanthus interruptus; G. coccinea – Graphocephala coccinea; H. bipunctulatus – Hindoloides bipunctulatus; H. coagulata – Homalodisca coagulata; H. communis – Helochara communis; H. elongata – Homalodisca elongata; H. oregonensis – Hylaius oregonensis; J. olitoria – Jikradia olitoria; L. coleoptrata – Lepyronia coleoptrata; M. costaricensis – Mahanarva costaricensis; M. sexnotatus – Macrosteles sexnotatus; M. striifrons – Macrosteles striifrons; M. septendecim – Magicicada septendecim; M. hiroglyphicus – Matsumuratettix hiroglyphicus; N. cincticeps – Nephotettix cincticeps; P. ferruginosa – Philya ferruginosa; P. grossus – Ptyelus grossus; P. modesta – Publilia modesta; P. reticulata – Pectinariophyes reticulata; P. spatulata – Pamplona spatulata; P. tredecimpunctata – Pagaronia tredecimpunctata; P. vittata – Platycotis vittata; S. festinus – Spisistilus festinus; T. garricola – Tibicina garricola; T. mariae – Tettigetta mariae)
The phylogeny of proteobacterial symbionts from the extant Cicadomorpha: Clypeata based on Bayesian analysis of partial 16S rDNA sequences (abbreviations for generic names as in Fig. 1). Dated tree generated using an uncorrelated longnormal clock in Beast. Posterior probabilities are indicated above the branches, whereas the numbers below the branches show the time of lineages divergence, scale is given with absolute geological ages (million years ago, MYA) (GenBank accession numbers of 16S rRNA genes are given in brackets)
Sequence divergences within Sulcia symbiont isolated from representatives of Cicadellidae range from 0.1 to 5 % (Table 1), whereas within the betaproteobacterial symbiont from 0.3 to 48.2 % (Table 2).
Ultrastructure, distribution and transovarial transmission of symbionts
The ovaries of Evacanthus interruptus are accompanied by large, paired, yellow-colored bacteriomes that are composed of two well-defined parts (Fig. 3a): (1) the part located in the close neighborhood to the body wall that is surrounded by a single layer of epithelial cells (bacteriome sheath) and (2) the part that adheres to the ovaries. Both parts of the bacteriomes consist of numerous giant bacteriocytes (Fig. 3a) which are tightly packed with morphologically distinct bacteria (Fig. 3b, c). Ultrastructural observations revealed that bacteriocytes occupying part of the bacteriome adhering to the body wall contain large, electron-dense, pleomorphic bacteria (Fig. 3b), whereas in bacteriocytes adhering to the ovaries there are large, pleomorphic, electron-translucent bacteria (Fig. 3c). The cytoplasm of the latter is more heterogenous (Fig. 3b) than the cytoplasm of the electron–dense bacteria (Fig. 3c). Both bacteria are characterized by the presence of electron-dense granules in their cytoplasm (Figs. 3b, c, 4c, g and 5c, d). The comparison of the results of morphological observations and results of molecular identification using specific pairs of primers amplifying 16S rRNA gene indicated that electron–dense, pleomorphic bacteria belong to the Bacteroidetes bacterium Sulcia, whereas electron–translucent bacteria represent the betaproteobacterial symbiont. Ultrastructural studies have not revealed the presence of bacteria in the bacteriome sheath.
Organization of the bacteriomes of Evacanthus interruptus. a Fragment of the bacteriome composed of bacteriocytes containing the bacterium Sulcia (bc1) and bacteriocytes containing betaproteobacterial symbiont (bc2). Bacteriocyte nucleus (bn), bacteriome sheath (bs). Methylene blue, scale bar = 20 μm. b Fragment of the bacteriocyte containing the bacterium Sulcia (S). Bacteriocyte nucleus (bn), electron-dense granule in the cytoplasm of Sulcia (in frame). TEM, scale bar = 2 μm. c Fragment of the bacteriocyte containing the betaproteobacterial symbiont (B). Bacteriocyte nucleus (bn), electron-dense granule in the cytoplasm of the betaproteobacterial symbiont (in frame). TEM, scale bar = 2 μm
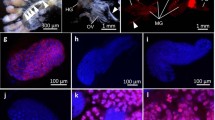
Transovarial transmission of symbionts. a, b Symbiotic bacteria (Sulcia - white arrows, betaproteobacterial symbionts - black arrows) that gather around the posterior pole of the terminal oocyte (oc) and start to enter cytoplasm of follicular cells (fc). Projection of the follicular cell (black asterisk), nucleus of follicular cell (fn). Methylene blue, scale bar = 20 μm. c Betaproteobacterial symbiont (B) enters (double black arrow) the projection (black asterisk) of the follicular cell (fc). Electron-dense granule in the cytoplasm of the betaproteobacterial symbiont (in frame). TEM, scale bar = 2 μm. d Fragment of the terminal oocyte (oc) (longitudinal section). Note bacteria Sulcia (white arrows) and betaproteobacterial symbionts (black arrows) that via massive projection (black asterisk) enter cytoplasm of follicular cells (fc). Nucleus of follicular cell (fn). Methylene blue, scale bar = 20 μm. e Fragment of the terminal oocyte (oc) (longitudinal section). Note enlarged follicular cells (fc) filled with symbiotic bacteria (encircled with dotted line) and symbiotic bacteria (Sulcia - white arrows, betaproteobacterial symbionts - black arrows) that invade follicular cells. Nucleus of follicular cell (fn). Methylene blue, scale bar = 20 μm. f Fragment of the terminal oocyte (oc) surrounded by follicular cells (fc) that are tightly packed by symbiotic bacteria (Sulcia - white arrows, betaproteobacterial symbionts - black arrows) (cross section). Nucleus of follicular cell (fn). Methylene blue, scale bar = 20 μm. g Bacteria Sulcia (S) and betaproteobacterial symbionts (B) in the cytoplasm of follicular cell. Nucleus of follicular cell (fn), electron-dense granules in the cytoplasm of the symbionts (in frames). TEM, scale bar = 2 μm
Transovarial transmission of symbionts. a Fragment of the terminal oocyte (oc) (longitudinal section). Note the follicular cells (fc) filled with symbiotic bacteria (Sulcia - white arrows, betaproteobacterial symbionts - black arrows) and accumulation of symbionts in the perivitelline space (white asterisk). Nucleus of follicular cell (fn), ovariolar stalk (pedicel) connecting the ovariole to the lateral oviduct (p), oolemma (arrowhead). Methylene blue, scale bar = 20 μm. b Fragment of the terminal oocyte during the late stage of infectioning by bacteria (cross section). Note the projections of follicular cells (fc) invaded by bacteria (Sulcia - white arrows, betaproteobacterial symbionts - black arrows) and the accumulation of symbionts in the perivitelline space (white asterisk). Nucleus of follicular cell (fn). Methylene blue, scale bar = 20 μm. c and d The accumulation of symbionts in the perivitelline space. Bacterium Sulcia (S), betaproteobacterial symbionts (B), oocyte (oc), oolemma (arrowhead), electron-dense granules in the cytoplasm of symbionts (in frames). TEM, scale bar= 2 μm. e The posterior pole of the oocyte (oc) (cross section). Note a characteristic ‘symbiont ball’(white asterisk) in the deep invagination of the oolemma (arrowhead). Follicular cells (fc), nucleus of follicular cell (fn), egg envelope (e). Fluorescence, confocal microscope, DAPI staining, scale bar = 20 μm
The symbionts are transmitted between generations transovarially, i.e. by means of the infection of oocytes. In the mature females which possess terminal oocytes at the stage of advanced vitellogenesis (for the characteristics of ovaries and process of oogenesis of Cicadomorpha see Książkiewicz-Kapralska 1985), both types of bacteria leave the cytoplasm of the bacteriocytes and begin to gather around the terminal oocytes (Fig. 4a, b, d). Then, the symbionts enter the cytoplasm of follicular cells which form a single-layered epithelium around the oocytes (Fig. 4a, b, d, e). During the bacterial invasion, the follicular cells surrounding the posterior pole of each terminal oocyte form massive projections (Fig. 4a, c, d) which enable the entry of these microorganisms into their cytoplasm. As the bacteria gradually accumulate into the cytoplasm, the volume of follicular cells markedly increases (Fig. 4e, f). As a result, the cytoplasm of the follicular cells is tightly packed with symbiotic microorganisms of both types (Fig. 4f, g). Next, the symbionts are gradually released into the perivitelline space (i.e. the space between the oocyte and follicular epithelium), where they assemble in the deep depression of the oolemma (Fig. 5a, b), forming finally a characteristic ‘symbiont ball’ (Fig. 5c, e). Up until the end of the oogenesis process (i.e. oocyte growth) the bacteria do not invade the ooplasm (i.e. they are isolated from the ooplasm by oolemma) (Fig. 5d, e). When the formation of the ‘symbiont ball’ has been finalized, the follicular cells surrounding the posterior pole of the oocyte begin synthesizing with the egg envelopes (Fig. 5e).
Discussion
Our observations revealed that two types of symbiotic bacteria are harbored in the body of Evacanthus interruptus (Evacanthinae: Evacanthini): Bacteroidetes symbionts Sulcia and betaproteobacterial symbiont closely related to the symbiont found in two other representatives of Cicadellidae: Pagaronia tredecimpunctata (Evacanthinae: Pagaronini) and Hylaius oregonensis (Bathysmatophorinae: Bathysmatophorini). These findings could suggest closer or more ancient relationships of these subfamilies. Dietrich (1999) considered Pagaronini and Evacanthini to be closely related, later (Dietrich 2004) united them together with Nirvanini in the subfamily Evacanthinae. However, available data (Dietrich 1999; Dietrich et al. 2001, 2005), provide no evidence of a close relationship of Evacanthinae to Bathysmatophorinae (Dietrich 2004; Wei et al. 2010). Known fossils cannot elucidate that relationship – the oldest Evacanthinae (Nirvanini) were reported from the Miocene Dominican amber (Dietrich and Vega 1995), while Bathysmatophorinae are more ancient, with fossil record reaching the late Cretaceous period (Szwedo 2005). The presence of the Bacteroidetes symbiont Sulcia and betaproteobacterial symbiont seems to be a very ancient and conservative heritage among Cicadellidae.
On the other hand, Evacanthus interruptus, Pagaronia tredecimpunctata and Hylaius oregonensis retained the ancestral betaproteobacterial symbiont which forms a well-defined clade with betaproteobacterial symbionts living in the members of the Deltocephalinae leafhoppers (see Fig. 2). These findings are consistent with the conclusion reached from the molecular data analysis indicating that betaproteobacterial symbionts are descendants of a bacterium which infected the shared ancestor of leafhoppers and spittlebugs (Bennett and Moran 2013; Koga et al. 2013; Koga and Moran 2014; Bennett et al. 2014). Recent molecular studies have also revealed that during the evolution of some lineages of the Cicadomorpha: Clypeata, the betaproteobacterium has been replaced by other bacteria (e.g. Baumannia, Hodgkinia, Sodalis-like bacteria – see Introduction). This more recent infection could be as ancient as the separation time of Cicadomorpha: Clypeata evolutionary lineage in the early Triassic period from the other Cicadomorpha (ancestral Hylicelloidea, and not directly related Palaeontinoidea and Dysmorphoptiloidea), which retained ancestral phloem-feeding (Wang et al. 2012), or even older. On the other hand, Shcherbakov (2012) stated that ‘leafhopperization’, i.e. the successive acquisition of membracoid characters leading to the modern Cicadellidae lineage, was a long process that spanned about a hundred million years, during the Jurassic and Cretaceous periods. Modern Cicadellidae diversity is definitively the result of their most recent radiation and, as for numerous other phytophagous groups, the effect of the Mid-Cretaceous biotic crisis and reorganization of the biosphere and later Cenozoic habitat diversification (Szwedo 2008). The oldest Euhemiptera (a unit comprising of Fulgoromorpha, Cicadomorpha, Coleorrhyncha and Heteroptera) have been reported from Late Carboniferous (Nel et al. 2013), so the infection with Bacteroidetes symbionts Sulcia and the betaproteobacterial symbiont could be more ancient (lost in Coleorrhyncha and Heteroptera). It took place probably more than 300 million years ago, maybe in relation to late Carboniferous rainforest collapse (DiMichele et al. 2010; Sahney et al. 2010; Cascales-Miñana and Cleal 2013).
Our studies did not revealed the presence in the body of E. interrupus of bacterium Baumannia that is characteristic for most members of Cicadellidae: Cicadellinae s. str. (Moran et al. 2003; Takiya et al. 2006; Wu et al. 2006; Bennett et al. 2014). This absence of Baumannia supports placement of Evacanthini in separate subfamily (Metcalf 1963; Dietrich 2004), and more remote relationships with Cicadellinae s. str., to which the tribe Evacanthini was formerly subordinated (Hamilton 1983; Oman et al. 1990), where Cicadellinae was treated sensu lato.
Earlier and more recent observations (Buchner 1925, 1965; Cheung and Purcell 1999; Noda et al. 2012; this study) indicate that of all of the leafhoppers examined thus far, the obligate symbionts are localized in the cytoplasm of separate bacteriocytes which are integrated into large organs termed bacteriomes. It seemed, thus, that in spite of the presence of diverse types of symbionts, the distribution of symbionts in the body of leafhoppers is similar. However, what has recently appeared is that in the green leafhopper, Cicadella viridis, its symbionts (i.e. Sulcia and Sodalis-like symbionts) may occur in separate bacteriocytes or may coexist in the same bacteriocyte (Michalik et al. 2014a). Moreover, it has been observed that in the common bacteriocytes Sodalis-like symbionts invade the cells of Sulcia, resulting in the occurrence in the cytoplasm of these bacteriocytes bacterium Sulcia and Sodalis-like symbionts inside the cells of Sulcia. Based on this observation, Michalik and co-workers (2014a) hypothesized that the symbiosis in Cicadella viridis represents the youngest state of association (i.e. the beginning of acquisition of Sodalis-like symbionts by Sulcia). In turn, in Macrosteles laevis (Kobiałka et al. 2014), some cells of Sulcia contain small rod-shaped bacteria, but the latter never occur individually. Thus, it seems that the distribution of symbionts and the state of symbiosis in members of the Cicadellidae is more complex than previously supposed.
Both earlier observations with the use of light microscopy (Buchner 1925, 1965; Müller 1962) and more recent ultrastructural studies (e.g. Cheung and Purcell 1999; Szklarzewicz and Moskal 2001; Szklarzewicz et al. 2006, 2013; Sacchi et al. 2008; Michalik et al. 2013; Michalik et al. 2014a, b; Kobiałka et al. 2014; Swiatoniowska et al. 2013; this study) have shown that in hemipterans, mycetomic symbionts may be transmitted from mother to the offspring in a different way, e.g. they may invade larval ovaries with undifferentiated germ cells (in some scale insects) or oocytes in adult females (in most hemipterans), they may be released from bacteriocytes and individually invade ovaries (in most hemipterans) or whole intact bacteriocytes may enter ovaries (in whiteflies). It should be stressed that even in closely related groups of hemipterans (e.g. within some families of scale insects), the transmission of symbionts may take a different course. Both earlier and recent studies (Buchner 1925, 1965; Müller 1962; Cheung and Purcell 1999; Sacchi et al. 2008; Michalik et al. 2014a; Kobiałka et al. 2014; this study) indicate that in Cicadomorpha, the symbionts infect the ovaries via follicular cells surrounding the posterior pole of the oocyte. Thus, in contrast to other hemipterans, all the lineages of Cicadomorpha: Clypeata developed the similar mechanism of transmission of symbionts from one generation to the next.
References
Andersen PC, Brodbeck BV, Mizell RF III (1989) Metabolism of amino acids, organic acids and sugars extracted from the xylem fluid of four host plants by adult Homalodisca coagulate. Entomol Exp Appl 50:149–159
Backus EA (1988) Sensory systems and behaviors which mediate hemipteran plant-feeding: a taxonomic overview. J Insect Physiol 34:151–157
Baumann P (2005) Biology of bacteriocyte-associated endosymbionts of plant sup-sucking insects. Annu Rev Microbiol 59:155–189
Bennett GM, Moran NA (2013) Small, smaller, smallest: the origin and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol 5:1675–1688
Bennett GM, McCutcheon JP, McDonald BR, Romanovicz D, Moran NA (2014) Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. mBio 5:e01697–14
Bressan A, Arneodo J, Simonato M, Haines WP, Boudon–Padieu E (2009) Characterization and evolution of two bacteriome-inhabiting symbionts in cixiid planthoppers (Hemiptera: Fulgoromorpha: Pentastirini). Environ Microbiol 11:3265–3279
Buchner P (1925) Studien an intracellularen Symbionten. V. Die symbiotischen Einrichtungen der Zikaden. Z Morphol Okol Tiere 4:88–245
Buchner P (1965) Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York
Campbell BC, Steffen-Campbell JD, Gill RJ (1994) Evolutionary origin of whiteflies (Hemiptera: Sternorrhyncha: Aleyrodidae) inferred from 18S rDNA sequences. Insect Mol Biol 3:73–88
Cascales-Miñana B, Cleal CJ (2013) The plant fossil record reflects just two great extinction events. Terra Nov 26:195–200
Cheung W, Purcell AH (1999) Invasion of bacteroids and BEV bacterium into oocytes of the leafhopper Euscelidius variegatus Kirschbaum (Homoptera: Cicadellidae): an electron microscopic study. Zool Stud 38:69–75
Dietrich CH (1999) The role of grasslands in the diversification of leafhoppers (Homoptera: Cicadellidae): a phylogenetic perspective. In: Warwick C (ed) Proceedings of the 15th North American Prairie Conference, St Charles, USA, pp 44–49
Dietrich CH (2004) Phylogeny of the leafhopper subfamily Evacanthinae with a review of neotropical species and notes on related groups (Hemiptera: Membracoidea: Cicadellidae). Syst Entomol 29:455–487
Dietrich CH (2013) Overview of the phylogeny, taxonomy and diversity of the leafhopper (Hemiptera: Auchenorrhyncha: Cicadomorpha: Membracoidea: Cicadellidae) vectors of plant pathogens. In: Chang C-J, Lee C-Y, Hsien-Tzung Shih H-T (eds) Proceedings of the 2013 International Symposium on Insect Vectors and Insect-Borne Diseases, Taichung, Taiwan, ROC. Special Publication of TARI 173:47–70
Dietrich CH, Vega FE (1995) Leafhoppers (Homoptera: Cicadellidae) from Dominican amber. Ann Entomol Soc Am 88:236–270
Dietrich CH, Rakitov RA, Holmes JL, Black WC (2001) Phylogeny of the major lineages of Membracoidea (Insecta: Hemiptera: Cicadomorpha) based on 28S rDNA sequences. Mol Phylogenet Evol 18:293–305
Dietrich CH, Dmitriev DA, Rakitov RA, Takiya DM, Zahniser JN (2005) Phylogeny of Cicadellidae (Cicadomorpha: Membracoidea) based on combined morphological and 28S rDNA sequence data. In: Purcell AH (ed) Abstracts of Talks and Posters, 12th International Auchenorrhyncha Congress, Berkeley, USA, pp S13–14
DiMichele WA, Cecil CB, Montañez IP, Falcon-Lang HJ (2010) Cyclic changes in Pennsylvanian paleoclimate and it effects on floristic dynamics in tropical Pangaea. Int J Coal Geol 8:329–344
Douglas AE (2006) Phloem sap feeding by animals: problems and solutions. J Exp Bot 57:747–754
Douglas AE (2009) The microbial dimension in insect nutritional ecology. Funct Ecol 23:38–47
Drohojowska J, Szwedo J, Azar D (2013) Talaya batraba gen. et sp. nov. - the first nymph of Protopsyllidiidae (Hemiptera: Sternorrhyncha: Psyllomorpha) from the Lower Cretaceous Lebanese amber. Acta Geol Sin Engl Ed 87:21–31
Forero D (2008) The systematics of the Hemiptera. Rev Colomb Entomol 34:1–21
Gonella E, Negri I, Marzorati M, Mandrioli M, Sacchi L, Pajoro M, Crotti E, Rizzi A, Clementi E, Tedeschi R, Bandi C, Alma A, Daffonchio D (2011) Bacterial endosymbiont localization in Hyalesthes obsoletus, the insect vector of Bois noir in Vitis vinifera. Appl Environ Microbiol 77:1423–1435
Grimaldi D, Engel MS (2005) Evolution of the insects. Cambridge University Press, Cambridge, 772 p
Hall TA (1999) BIOEDIT: an user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp 41:95–98
Hamilton KGA (1983) Classification, morphology and phylogeny of the family Cicadellidae (Rhynchota, Homoptera). In: Knight WJ, Pant NC, Robertson TS, Wilson MR (eds) Proc 1st Int Wksp Biotax, Classif, Biol Leafhoppers Planthoppers Econ Impt, London, UK, 4–7 October 1982. CIE, London, pp 15–37
Ishii Y, Matsuura Y, Kakizawa S, Nikoh N, Fukatsu T (2013) Diversity of bacterial endosymbionts associated with Macrosteles leafhoppers vectoring phytopathogenic phytoplasmas. Appl Environ Microbiol 79:5013–5022
Kikuchi Y (2009) Endosymbiotic bacteria in insects: their diversity and culturability. Microbes Environ 24:195–204
Kobiałka M, Michalik A, Szklarzewicz T (2014) Endosymbiotic bacteria in Macrosteles laevis (Insecta, Hemiptera, Cicadellidae: Deltocephalinae). Ultrastructure, distribution and transovarial transmission. Acta Biol Cracov Ser Bot 56(suppl 1):66
Koga R, Moran NA (2014) Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J 8:1237–1246
Koga R, Bennett GM, Cryan JR, Moran NA (2013) Evolutionary replacement of symbionts in an ancient and diverse insect lineage. Environ Microbiol 15:2073–2081
Książkiewicz-Kapralska M (1985) Organization of the trophic chamber of homopteran insects. 1. Cercopidae: Cicadomorpha. Cytobios 42:133–145
McCutcheon JP, Moran NA (2007) Parallel genomic evolution and metabolic interdependense in an ancient symbiosis. Proc Natl Acad Sci U S A 104:19392–19397
McCutcheon JP, McDonald BR, Moran NA (2009) Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A 106:15394–15399
Metcalf ZP (1963) General Catalogue of the Homoptera, Fascicle VI Cicadelloidea, Part 6 Evacanthidae. US DeptAgric, ARS, Washington, DC
Michalik A, Jankowska W, Szklarzewicz T (2009) Ultrastructure and transovarial transmission of endosymbiotic microorgansms in Conomelus anceps and Metcalfa pruinosa (Insecta, Hemiptera, Fulgoromorpha). Folia Biol (Kraków) 57:131–137
Michalik A, Gołas A, Kot M, Wieczorek K, Szklarzewicz T (2013) Endosymbiotic microorganisms in Adelges (Sacchiphantes) viridis (Insecta, Hemiptera, Adelgoidea: Adelgidae): molecular characterization, ultrastructure and transovarial transmission. Arthropod Struct Dev 42:531–538
Michalik A, Jankowska W, Kot M, Gołas A, Szklarzewicz T (2014a) Symbiosis in the green leafhopper, Cicadella viridis (Hemiptera, Cicadellidae). Association in statu nascendi? Arthropod Struct Dev 43:579–587
Michalik A, Szklarzewicz T, Jankowska W, Wieczorek K (2014b) Endosymbiotic microorganisms of aphids (Hemiptera: Sternorrhyncha: Aphidoidea): ultrastructure, distribution and transovarial transmission. Eur J Entomol 111:91–104
Moran NA, Dale C, Dunbar H, Smith W, Ochman H (2003) Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ Microbiol 5:116–126
Moran NA, Tran P, Gerardo NM (2005) Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol 71:8802–8810
Müller HJ (1962) Neuere Vorstellungen über Verbreitung und Phylogenie der Endosymbiosen der Zikaden. Z Morphol Okol Tiere 51:190–210
Nel A, Roques P, Nel P, Prokin AA, Bourgoin T, Prokop J, Szwedo J, Azar D, Desutter-Grandcolas L, Wappler T, Garrouste R, Coty D, Huang D-Y, Engel MS, Kirejtshuk AG (2013) The earliest known holometabolous insects. Nature 503:257–261
Noda H (1977) Histological and histochemical observation of intracellular yeast–like symbiotes in the fat body of the small brown planthopper, Laodelphax striatellus (Homoptera: Delphacidae). Appl Entomol Zool 12:134–141
Noda H, Watanabe K, Kawai S, Yukuhiro F, Miyoshi T, Tomizawa M, Koizumi Y, Nikoh N, Fukatsu T (2012) Bacteriome-associated endosymbionts of the green rice leafhopper Nephotettix cincticeps (Hemiptera: Cicadellidae). Appl Entomol Zool 47:217–225
Oliver KM, Russel JA, Moran NA, Hunter MS (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100:1803–1807
Oman PW, Knight WJ, Nielson MW (1990) Leafhoppers (Cicadellidae): a bibliography, generic check-list, and index to the world literature, 1956–1985. CAB Int Inst Ent, Wallingford
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 1:817–818
Rambaut A (2008) FigTree v. 1.4.0. Available http://tree.bio.ed.ac.uk/software/figtree/. Accessed 20 May 2013. Rambaut
Rambaut A, Drummond AJ (2007) Tracer v1.4: MCMC trace analyses tool. Available: http://beast.bio.ed.ac.uk/Tracer
Sacchi L, Genchi M, Clementi E, Bigliardi E, Avanzatti AM, Pajoroi M, Negri I, Marzorati M, Gonella E, Alma A, Daffonchio D, Bandi C (2008) Multiple symbiosis in the leafhopper Scaphoideus titanus (Hemiptera: Cicadellidae): details of transovarial transmission of Cardinium sp. and yeast-like endosymbionts. Tissue Cell 40:231–242
Sahney S, Benton MJ, Falcon-Lang HJ (2010) Rainforest collapse triggered Pennsylvanian tetrapod diversification in Euramerica. Geology 38:1079–1082
Scarborough CL, Ferrari J, Godfray HCJ (2005) Aphid protected from pathogen by endosymbiont. Science 310:1781
Shcherbakov DE (2012) More on Mesozoic Membracoidea (Homoptera). Russ Entomol J 21:15–22
Sorensen JT, Cambell BC, Gill RJ, Steffen-Campbell JD (1995) Non-monophyly of Auchenorrhyncha (‘Homoptera’), based upon 18S rDNA phylogeny: ecoevolutionary and cladistic implications within pre-Heteropterodea Hemiptera (s.l.) and a proposal for new monophyletic suborders. Pan Pac Entomol 71:31–60
Swiatoniowska M, Ogorzalek A, Golas A, Michalik A, Szklarzewicz T (2013) Ultrastructure, distribution and transovarial transmission of symbiotic microorganisms in Nysius ericae and Nithecus jacobaeae (Heteroptera: Lygaeidae: Orsillinae). Protoplasma 250:325–332
Szklarzewicz T, Moskal A (2001) Ultrastructure, distribution, and transmission of endosymbionts in the whitefly Aleurochiton aceris Modeer (Insecta, Hemiptera, Aleyrodinea). Protoplasma 218:45–53
Szklarzewicz T, Kędra K, Niżnik S (2006) Ultrastructure and transovarial transmission of endosymbiotic microorganisms in Palaeococcus fuscipennis (Burmeister) (Insecta, Hemiptera, Coccinea: Monophlebidae). Folia Biol (Kraków) 54:69–74
Szklarzewicz T, Kalandyk-Kolodziejczyk M, Kot M, Michalik A (2013) Ovary structure and transovarial transmission of endosymbiotic microorganisms in Marchalina hellenica (Insecta, Hemiptera, Coccomorpha: Marchalinidae). Acta Zool (Stockholm) 94:184–192
Szwedo J (2005) Jantarivacanthus kotejai gen. et sp. n. from Eocene Baltic amber, with notes on the Bathysmatophorini and related taxa (Hemiptera: Cicadomorpha: Cicadellidae). Pol J Entomol 74:251–276
Szwedo J (2008) Paradise Lost? - Cretaceous and Palaeogene diversification of planthoppers and leafhoppers (Hemiptera: Fulgoromorpha et Cicadomorpha). Bull Insectol 61:111–112
Szwedo J, Bourgoin T, Lefebvre F (2004) Fossil planthoppers (Hemiptera: Fulgoromorpha) of the world. An annotated catalogue with notes on Hemiptera classification. Studio 1, Warszawa. 208 p
Takiya DM, Tran P, Dietrich CH, Moran NA (2006) Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol Ecol 15:4175–4191
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Urban J, Cryan J (2012) Two ancient bacterial endosymbionts have coevolved with the planthoppers (Insecta: Hemiptera: Fulgoroidea). BMC Evol Biol 12:87
Wang B, Szwedo J, Zhang H-C (2012) New Cercopoidea from the Middle Jurassic of China and their evolutionary significance (Insecta: Hemiptera: Cicadomorpha). Palaeontology 55:1223–1243
Wei C, Zhang Y, Dietrich CH (2010) A new brachypterous leafhopper of the tribe Malmaemichungiini (Hemiptera: Cicadellidae: Bathysmatophorinae), representing the first record of the tribe from China. Zootaxa 2689:48–56
Wilkinson TL, Ishikawa H (2001) On the functional significance of symbiotic microorganisms in the Homoptera: a comparative study of Acyrthosiphon pisum and Nilaparvata lugens. Physiol Entomol 26:86–93
Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H (2006) Metabolic complementarity and genomics of the dual symbiosis of sharpshooters. PLoS Biol 4, e188
Acknowledgments
We are greatly indebted to Prof. Józefa Styrna (Department of Genetics and Evolution, Jagiellonian University, Kraków) for providing the equipment for molecular analyses and Ada Jankowska and Olga Woźnicka for their skilled technical assistance.
This work was supported by funds from research grant K/ZDS/004838 (Jagiellonian University).
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Hanns H. Kassemeyer
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Szklarzewicz, T., Grzywacz, B., Szwedo, J. et al. Bacterial symbionts of the leafhopper Evacanthus interruptus (Linnaeus, 1758) (Insecta, Hemiptera, Cicadellidae: Evacanthinae). Protoplasma 253, 379–391 (2016). https://doi.org/10.1007/s00709-015-0817-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-015-0817-2