Abstract
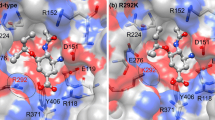
The recent H1N1 influenza pandemic has attracted worldwide attention due to the high infection rate. Oseltamivir is a new class of anti-viral agent approved for the treatment and prevention of influenza infections. The principal target for this drug is a virus surface glycoprotein, neuraminidase (NA), which facilitates the release of nascent virus and thus spreads infection. Until recently, only a low prevalence of neuraminidase inhibitor (NAI) resistance (<1 %) had been detected in circulating viruses. However, there have been reports of significant numbers of A (H1N1) influenza strains with a N294S neuraminidase mutation that was highly resistant to the NAI, oseltamivir. Hence, in the present study, we highlight the effect of point mutation-induced oseltamivir resistance in H1N1 subtype neuraminidases by molecular simulation approach. The docking analysis reveals that mutation (N294S) significantly affects the binding affinity of oseltamivir with mutant type NA. This is mainly due to the decrease in the flexibility of binding site residues and the difference in prevalence of hydrogen bonds in the wild and mutant structures. This study throws light on the possible effects of drug-resistant mutations on the large functionally important collective motions in biological systems.













Similar content being viewed by others
References
Alexandrov V, Lehnert U, Echols N, Milburn D, Engelman D, Gerstein M (2005) Normal modes for predicting protein motions: a comprehensive database assessment and associated Web tool. Protein Sci 14(3):633–643
Alves ID, Bechara C, Walrant A, Zaltsman Y, Jiao CY, Sagan S (2011) Relationships between membrane binding, affinity and cell internalization efficacy of a cell-penetrating peptide: penetratin as a case study. PLoS One 6(9):e24096
Baum SG (2009) Oseltamivir resistance: what does it mean clinically? Clin Infect Dis 49(12):1836–1837
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28(1):235–242
Bucher DJ, Kilbourne ED (1972) A 2 (N2) neuraminidase of the X-7 influenza virus recombinant: determination of molecular size and subunit composition of the active unit. J Virol 10(1):60–66
Carlson HA, McCammon JA (2000) Accommodating protein flexibility in computational drug design. Mol Pharmacol 57(2):213–218
Cavasotto CN, Kovacs JA, Abagyan RA (2005) Representing receptor flexibility in ligand docking through relevant normal modes. J Am Chem Soc 127(26):9632–9640
Choudhury D, Biswas S, Roy S, Dattagupta JK (2010) Improving thermostability of papain through structure-based protein engineering. Protein Eng Des Sel 23(6):457–467
Collins PJ, Haire LF, Lin YP, Liu J, Russell RJ, Walker PA, Skehel JJ, Martin SR, Hay AJ, Gamblin SJ (2008) Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 453(7199):1258–1261
Colman PM, Varghese JN, Laver WG (1983) Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303(5912):41–44
Darden T, Perera L, Li L, Pedersen L (1999) New tricks for modelers from the crystallography toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations. Structure 7(3):55–60
Delarue M, Dumas P (2004) On the use of low-frequency normal modes to enforce collective movements in refining macromolecular structural models. Proc Natl Acad Sci U S A 101(18):6957–62
Du QS, Wang SQ, Chou KC (2007) Study of drug resistance of chicken influenza A virus (H5N1) from homology-modeled 3D structures of neuraminidases. Biochem Biophys Res Commun 354(3):634–640
Feldman J, Snyder KA, Ticoll A, Pintilie G, Hogue CW (2006) A complete small molecule dataset from the protein data bank. FEBS Lett 580(6):1649–1653
Gasteiger J, Rudolph C, Sadowski J (1990) Automatic generation of 3D-atomic coordinates for organic molecules. Tetrahedron Comput Meth 3:537–547
Gubareva LV, Kaiser L, Hayden FG (2000) Influenza virus neuraminidase inhibitors. Lancet 355(9206):827–835
Hess B, Kutzner C, Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4(3):435–447
Hinkle A, Tobacman LS (2003) Folding and function of the troponin tail domain. Effects of cardiomyopathic troponin T mutations. J Biol Chem 278(1):506–513
Hsieh YC, Wu TZ, Liu DP, Shao PL, Chang LY, Lu CY, Lee CY, Huang FY, Huang LM (2006) Influenza pandemics: past, present and future. J F Med Assoc 105(1):1–6
Kim CU, Lew W, Williams MA, Liu H, Zhang L, Swaminathan S, Bischofberger N, Chen MS, Mendel DB, Tai CY (1997) Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J Am Chem Soc 119(4):681–690
Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson J (2008) Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure 16(9):1399–406
Lindahl E, Hess B, van der Spoel D (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Mod 7(8):306–317
Lopez G, Valencia A, Tress ML (2007) firestar—prediction of functionally important residues using structural templates and alignment reliability. Nucleic Acids Res 35:573–577
Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk H (2004) Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol 78(22):12665–12667
McKimm-Breschkin JL (2000) Resistance of influenza viruses to neuraminidase inhibitors—a review. Antiviral Res 47(1):1–17
Meagher KL, Carlson HA (2005) Solvation influences flap collapse in HIV-1 protease. Proteins: Struct Func Bioinf 58(1):119–125
Moscona A (2005) Oseltamivir resistance—disabling our influenza defenses. N Engl J Med 353(25):2633–2636
Mukhtar MM, Rasool ST, Song D, Zhu C, Hao Q, Zhu Y, Wu J (2007) Origin of highly pathogenic H5N1 avian influenza virus in China and genetic characterization of donor and recipient viruses. J Gen Virol 88(11):3094–3099
Pales PS (2004) Influenza: old and new threats. Nat Med 10:82–87
Parthasarathy S, Murthy MR (2000) Protein thermal stability: insights from atomic displacement parameters (B values). Protein Eng 13(1):9–13
Rajasekaran R, George Priya Doss C, Sudandiradoss C, Ramanathan K, Purohit R, Sethumadhavan R (2008) Effect of deleterious nsSNP on the HER2 receptor based on stability and binding affinity with herceptin: a computational approach. C R Biol 331(6):409–417
Ringe D, Petsko GA (1986) Study of protein dynamics by X-ray diffraction. Methods Enzymol 131:389–433
Rungrotmongkol T, Udommaneethanakit T, Malaisree M, Nunthaboot N, Intharathep P, Sompornpisut P, Hannongbua S (2009) How does each substituent functional group of oseltamivir lose its activity against virulent H5N1 influenza mutants? Biophys Chem 145(1):29–36
Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ (2006) The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443(7107):45–49
Shirvan AN, Moradi M, Aminian M, Madani R (2007) Preparation of neuraminidase-specific antiserum from the H9N2 subtype of avian influenza virus Turk. J Vet Anim 31(4):219–223
Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. Comput Chem 26(16):1701–1718
Su Y, Yang HY, Zhang BJ, Jia HL, Tien P (2008) Analysis of a point mutation in H5N1 avian influenza virus hemagglutinin in relation to virus entry into live mammalian cells. Arch Virol 153(12):2253–2261
Suhre K, Sanejouand YH (2004) ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids 32:610–614
Takeda M, Leser GP, Russell CJ, Lamb RA (2003) Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc Natl Acad Sci 100(25):14610–14617
Teague SJ (2003) Implications of protein flexibility for drug discovery. Nat Rev Drug Discov 2(7):527–541
Thompson MA (2004) ArgusLab 4.0.1. Planaria Software, Seattle. http://www.ArgusLab.com
Thompson MA (2004b) Poster presentation: molecular docking using arguslab: An efficient shape-based search algorithm and the AScore scoring function. Fall ACS meeting, Philadelphia
Varghese JN, Smith PW, Sollis SL, Blick SJ, Sahasrabudhe A, Mckimm-Breschkin JL, Colman PM (1998) Drug design against a shifting target: a structural basis for resistance to inhibitors in a variant of influenza virus neuraminidase. Structure 6(6):735–746
Von Itzstein M, Wu WY, Kok BG, Pegg MS, Dyason JC, Jin B, Van Phan T, Smythe ML, White HF, Oliver SW (1993) Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363(6428):418–423
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng 8(2):127–134
Yuan Z, Bailey TL, Teasdale RD (2005) Prediction of protein B-factor profiles. Proteins 58(4):905–912
Acknowledgments
The authors express deep sense of gratitude to the management of Vellore Institute of Technology for all the support, assistance, and constant encouragement to carry out this work. The authors also thank Professor M.A. Mohamed Sahul Hameed, English Division, for grammar corrections in our manuscript. The authors also thank reviewers for their valuable comments and suggestion in the improvement of our manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Peter Nick
Rights and permissions
About this article
Cite this article
Karthick, V., Shanthi, V., Rajasekaran, R. et al. In silico analysis of drug-resistant mutant of neuraminidase (N294S) against oseltamivir. Protoplasma 250, 197–207 (2013). https://doi.org/10.1007/s00709-012-0394-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-012-0394-6