Abstract
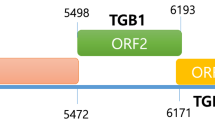
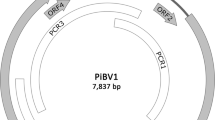
A novel betaflexivirus, tentatively named “miscanthus virus M” (MiVM), was isolated from Miscanthus sp. The complete genome of MiVM is 7,388 nt in length (excluding the poly(A) tail). It contains five open reading frames and has a genome organization similar to those of members of the families Alphaflexiviridae and Betaflexiviridae (subfamily Quinvirinae). The amino acid sequences of both the replicase and coat protein shared less than 45% identity with the corresponding sequences of members of either family. Phylogenetic analysis confirmed that MiVM belongs to the family Betaflexiviridae and subfamily Quinvirinae but it was too distantly related to be included in any currently recognized genus in this family. We therefore propose that miscanthus virus M represents a new species and a new genus in the family Betaflexiviridae.

Similar content being viewed by others
Data availability
The viral genomic sequence generated in this work has been deposited in the GenBank database of the National Center for Biotechnology Information (NCBI) under the accession number ON986335.
References
Lewandowski I, Clifton-Brown J, Kiesel A, Hastings A, Iqbal Y (2018) 2-Miscanthus. Perennial grasses for bioenergy and bioproducts: production, uses, sustainability and markets for giant reed, miscanthus, switchgrass, reed canary grass and bamboo. Academic Publications, pp 35–59. https://doi.org/10.1016/B978-0-12-812900-5.00002-3
Moll L, Wever C, Völkering G, Pude R (2020) Increase of Miscanthus cultivation with new roles in materials production—a review. Agronomy 10:308. https://doi.org/10.3390/agronomy10020308
Agindotan BO, Ahonsi MO, Domier LL, Gray ME, Bradley CA (2010) Application of sequence-independent amplification (SIA) for the identification of RNA viruses in bioenergy crops. J Virol Methods 169:119–128. https://doi.org/10.1016/j.jviromet.2010.07.008
Agindotan B, Okanu N, Oladeinde A, Voigt T, Long S, Gray M, Bradley C (2013) Detection of switchgrass mosaic virus in Miscanthus and other grasses. Can J Plant Pathol 35:81–86. https://doi.org/10.1080/07060661.2012.752763
Grisham MP, Maroon-Lango CJ, Hale AL (2012) First report of sorghum mosaic virus causing mosaic in Miscanthus sinensis. Plant Dis 96:150. https://doi.org/10.1094/PDIS-07-11-0617
Bolus S, Malapi-Wight M, Grinstead SC, Fuentes-Bueno I, Hendrickson L, Hammond RW, Mollov D (2020) Identification and characterization of Miscanthus yellow fleck virus, a new polerovirus infecting Miscanthus sinensis. PLoS ONE 15:e0239199. https://doi.org/10.1371/journal.pone.0239199
Costa LC, Hu X, Malapi-Wight M, O’Connell M, Hendrickson LM, Turner RS, McFarland C, Foster J, Hurtado-Gonzales OP (2022) Genomic characterization of silvergrass cryptic virus 1, a novel partitivirus infecting Miscanthus sinensis. Arch Virol 167:261–265. https://doi.org/10.1007/s00705-021-05294-6
Leblanc Z, Gauthier ME, Lelwala R, Elliott C, McMaster C, Eichner R, Davis K, Liefting L, Thompson J, Dinsdale A, Whattam M (2022) Complete genome sequence of a novel potyvirus infecting Miscanthus sinensis (silver grass). Arch Virol 167:1701–1705. https://doi.org/10.1007/s00705-022-05445-3
Chatani M, Matsumoto Y, Mizuta H, Ikegami M, Boulton MI, Davies JW (1991) The nucleotide sequence and genome structure of the geminivirus miscanthus streak virus. J Gen Virol 72:2325–2331. https://doi.org/10.1099/0022-1317-72-10-2325
Kar S, Zhang N, Nakashima T, Villanueva-Morales A, Stewart JR, Sacks EJ, Terajima Y, Yamada T (2019) Saccharum × Miscanthus intergeneric hybrids (miscanes) exhibit greater chilling tolerance of C4 photosynthesis and postchilling recovery than sugarcane (Saccharum spp. hybrids. GCB Bioenergy 11:1318–1333. https://doi.org/10.1111/gcbb.12632
Silva JM, Melo FL, Elena SF, Candresse T, Sabanadzovic S, Tzanetakis IE, Blouin AG, Villamor DE, Mollov D, Constable F, Cao M (2022) Virus classification based on in-depth sequence analyses and development of demarcation criteria using the Betaflexiviridae as a case study. J Gen Virol 103:001806. https://doi.org/10.1099/jgv.0.001806
Morozov SY, Solovyev AG (2020) Small hydrophobic viral proteins involved in intercellular movement of diverse plant virus genomes. AIMS Microbiol 6:305. https://doi.org/10.3934/microbiol.2020019
Atabekova AK, Solovieva AD, Chergintsev DA, Solovyev AG, Morozov SY (2023) Role of plant virus movement proteins in suppression of host RNAi defense. Int J Mol Sci 24:9049. https://doi.org/10.3390/ijms24109049
Adams MJ, Candresse T, Hammond J, Kreuze JF, Martelli GP, Mamba S, Pearson MN, Ryu KH, Saldarelli P, Yoshikawa N (2012) Betafexiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy, ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, Cambridge, pp 920–941
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9:e108277. https://doi.org/10.1371/journal.pone.0108277
Funding
This work was partly funded by an in-house-appropriated project (number 8042-22000-324-000-D).
Author information
Authors and Affiliations
Contributions
Conceptualization: DM. Formal analysis: PA, SG. Investigation: PA, SG, RG, PR, DM. Methodology: PA, SG, GRK, RG, PR, DM. Writing – original draft: PA, DM. Writing – review & editing: PA, SG, GRK, RG, PR, DM.
Corresponding authors
Ethics declarations
Ethical approval
This article does not contain any studies with animals or human participants performed by any of the authors.
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Jesús Navas-Castillo
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Abrahamian, P., Grinstead, S., Kinard, G.R. et al. Complete sequence and genome characterization of miscanthus virus M, a new betaflexivirus from Miscanthus sp.. Arch Virol 169, 27 (2024). https://doi.org/10.1007/s00705-024-05966-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-024-05966-z