Abstract
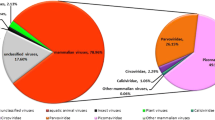
Coyotes (Canis latrans) have a broad geographic distribution across North and Central America. Despite their widespread presence in urban environments in the USA, there is limited information regarding viruses associated with coyotes in the USA and in particular the state of Arizona. To explore viruses associated with coyotes, particularly small DNA viruses, 44 scat samples were collected (April–June 2021 and November 2021–January 2022) along the Salt River near Phoenix, Arizona (USA), along 43 transects (500 m). From these samples, we identified 11 viral genomes: two novel circoviruses, six unclassified cressdnaviruses, and two anelloviruses. One of the circoviruses is most closely related to a circovirus sequence identified from an aerosolized dust sample in Arizona, USA. The second circovirus is most closely related to a rodent-associated circovirus and canine circovirus. Of the unclassified cressdnaviruses, three encode replication-associated proteins that are similar to those found in protists (Histomonas meleagridis and Monocercomonoides exilis), implying an evolutionary relationship with or a connection to similar unidentified protist hosts. The two anelloviruses are most closely related to those found in rodents, and this suggests a diet-related identification.






Similar content being viewed by others
Data availability
The sequences described in this study have been deposited in the GenBank database under accession numbers OQ599920-OQ599930.
References
Bateman PW, Fleming PA, Le Comber S (2012) Big city life: carnivores in urban environments. J Zool 287:1–23
Hody JW, Kays R (2018) Mapping the expansion of coyotes (Canis latrans) across North and Central America. Zookeys 759:81–97
Snow CJ (1967) Some observations on the behavioral and morphological development of coyote pups. Am Zool 7:353–355
Clark KA, Neill SU, Smith JS, Wilson PJ, Whadford VW, McKirahan GW (1994) Epizootic canine rabies transmitted by coyotes in south Texas. J Am Vet Med Assoc 204:536–540
Morey PS, Gese EM, Gehrt S (2007) Spatial and temporal variation in the diet of coyotes in the chicago metropolitan area. Am Midl Nat 158:147–161
Poessel SA, Mock EC, Breck SW (2017) Coyote (Canis latrans) diet in an urban environment: variation relative to pet conflicts, housing density, and season. Can J Zool 95:287–297
Sacks BN, Neale JCC (2002) Foraging strategy of a generalist predator toward a special prey: coyote predation on sheep. Ecol Appl 12:299–306
Ma X, Bonaparte S, Corbett P, Orciari LA, Gigante CM, Kirby JD, Chipman RB, Fehlner-Gardiner C, Thang C, Cedillo VG (2023) Rabies surveillance in the United States during 2021. J Am Vet Med Assoc 261:1045–1053
Krebs JW, Noll HR, Rupprecht CE, Childs JE (2002) Rabies surveillance in the United States during 2001. J Am Vet Med Assoc 221:1690–1701
Blanton JD, Robertson K, Palmer D, Rupprecht CE (2009) Rabies surveillance in the United States during 2008. J Am Vet Med Assoc 235:676–689
Malmlov A, Breck S, Fry T, Duncan C (2014) Serologic survey for cross-species pathogens in urban coyotes (Canis latrans), Colorado, USA. J Wildl Dis 50:946–950
Wang X, Brown CM, Smole S, Werner BG, Han L, Farris M, DeMaria A (2010) Aggression and rabid coyotes, Massachusetts, USA. Emerg Infect Dis 16:357–359
Velasco-Villa A, Orciari LA, Souza V, Juarez-Islas V, Gomez-Sierra M, Castillo A, Flisser A, Rupprecht CE (2005) Molecular epizootiology of rabies associated with terrestrial carnivores in Mexico. Virus Res 111:13–27
Gese EM, Schultz RD, Johnson MR, Williams ES, Crabtree RL, Ruff RL (1997) Serological survey for diseases in free-ranging coyotes (Canis latrans) in Yellowstone National Park, Wyoming. J Wildl Dis 33:47–56
Bischof R, Rogers DG (2005) Serologic survey of select infectious diseases in coyotes and raccoons in Nebraska. J Wildl Dis 41:787–791
Guo W, Evermann JF, Foreyt WJ, Knowlton FF, Windberg LA (1986) Canine distemper virus in coyotes: a serologic survey. J Am Vet Med Assoc 189:1099–1100
Allison AB, Kohler DJ, Ortega A, Hoover EA, Grove DM, Holmes EC, Parrish CR (2014) Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLoS Pathog 10:e1004475
Canuti M, Mira F, Sorensen RG, Rodrigues B, Bouchard E, Walzthoni N, Hopson M, Gilroy C, Whitney HG, Lang AS (2022) Distribution and diversity of dog parvoviruses in wild, free-roaming and domestic canids of Newfoundland and Labrador, Canada. Transbound Emerg Dis 69:e2694–e2705
Canuti M, Rodrigues B, Whitney HG, Lang AS (2017) Introduction of canine parvovirus 2 into wildlife on the Island of Newfoundland, Canada. Infect Genet Evol 55:205–208
Gese EM, Grothe S (1995) Analysis of coyote predation on deer and elk during winter in Yellowstone National Park, Wyoming. Am Midl Nat 133:36–43
Pluemer M, Dubay S, Drake D, Crimmins S, Veverka T, Hovanec H, Torkelson M, Mueller M (2019) Red foxes (Vulpes vulpes) and coyotes (Canis latrans) in an urban landscape: prevalence and risk factors for disease. J Urban Ecol. https://doi.org/10.1093/jue/juz022
Brown JL, Wold B, Larson RN (2020) LA urban coyote project volunteer training. https://doi.org/10.17504/protocols.io.933h8qn
Morin DJ, Higdon SD, Lonsinger RC, Gosselin EN, Kelly MJ, Waits LP (2019) Comparing methods of estimating carnivore diets with uncertainty and imperfect detection. Wildl Soc Bull 43:651–660
Debelica A, Thies ML (2009) Atlas and key to the hair of terrestrial Texas mammals. Museum of Texas Tech University, Lubbock
Larson RN, Brown JL, Karels T, Riley SP (2020) Effects of urbanization on resource use and individual specialization in coyotes (Canis latrans) in southern California. PLoS ONE 15:e0228881
Moore TD, Spence LE, Dugnolle CE (1997) Identification of the dorsal guard hairs of some mammals of Wyoming. Wyoming Game and Fish Department
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, Yamashita H, Lam TW (2016) MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102:3–11
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Zallot R, Oberg N, Gerlt JA (2019) The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 58:4169–4182
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534
Letunic I, Bork P (2021) Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296
Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Stover BC, Muller KF (2010) TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform 11:7
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9:e108277
Krupovic M, Varsani A, Kazlauskas D, Breitbart M, Delwart E, Rosario K, Yutin N, Wolf YI, Harrach B, Zerbini FM, Dolja VV, Kuhn JH, Koonin EV (2020) Cressdnaviricota: a virus phylum unifying seven families of rep-encoding viruses with single-stranded, circular DNA genomes. J Virol 94:e00582-00520
Ilyina TV, Koonin EV (1992) Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from Eubacteria, Eucaryotes and Archaebacteria. Nucleic Acids Res 20:3279–3285
Koonin EV, Ilyina TV (1993) Computer-assisted dissection of rolling circle DNA replication. Biosystems 30:241–268
Rosario K, Duffy S, Breitbart M (2012) A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol 157:1851–1871
Zhao L, Rosario K, Breitbart M, Duffy S (2019) Eukaryotic circular rep-encoding single-stranded DNA (CRESS DNA) viruses: ubiquitous viruses with small genomes and a diverse host range. Adv Virus Res 103:71–133
Kazlauskas D, Varsani A, Krupovic M (2018) Pervasive chimerism in the replication-associated proteins of uncultured single-stranded DNA viruses. Viruses 10:187
Kazlauskas D, Varsani A, Koonin EV, Krupovic M (2019) Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nat Commun 10:3425
Tisza MJ, Pastrana DV, Welch NL, Stewart B, Peretti A, Starrett GJ, Pang Y-YS, Krishnamurthy SR, Pesavento PA, McDermott DH, Murphy PM, Whited JL, Miller B, Brenchley J, Rosshart SP, Rehermann B, Doorbar J, Ta’ala BA, Pletnikova O, Troncoso JC, Resnick SM, Bolduc B, Sullivan MB, Varsani A, Segall AM, Buck CB (2020) Discovery of several thousand highly diverse circular DNA viruses. Elife 9:e51971
Kinsella CM, Deijs M, Becker C, Broekhuizen P, van Gool T, Bart A, Schaefer AS, van der Hoek L (2022) Host prediction for disease-associated gastrointestinal cressdnaviruses. Virus Evol 8:vea087
Chrzastek K, Kraberger S, Schmidlin K, Fontenele RS, Kulkarni A, Chappell L, Dufour-Zavala L, Kapczynski DR, Varsani A (2021) Diverse single-stranded DNA viruses identified in chicken buccal swabs. Microorganisms 9:2602
Harding C, Larsen BB, Otto HW, Potticary AL, Kraberger K, Custer JM, Suazo C, Upham NS, Worobey M, van Doorslaer K, Varsani A (2022) Diverse DNA virus genomes identified in fecal samples of Mexican free-tailed bats (Tadarida brasiliensis) captured in Chiricahua Mountains of southeast Arizona (USA). Virology 580:98–111
Lund MC, Larsen BB, Rowsey DM, Otto HW, Gryseels S, Kraberger S, Custer JM, Steger L, Yule KM, Harris RE, Worobey M, Van Doorslaer K, Upham NS, Varsani A (2023) Using archived and biocollection samples towards deciphering the DNA virus diversity associated with rodent species in the families Cricetidae and Heteromyidae. Virology 585:42–60
Rosario K, Breitbart M, Harrach B, Segales J, Delwart E, Biagini P, Varsani A (2017) Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol 162:1447–1463
Breitbart M, Delwart E, Rosario K, Segales J, Varsani A, Ictv Report C (2017) ICTV virus taxonomy profile: Circoviridae. J Gen Virol 98:1997–1998
Pass DA, Perry RA (1984) The pathology of psittacine beak and feather disease. Aust Vet J 61:69–74
Meng XJ (2013) Porcine circovirus type 2 (PCV2): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci 1:43–64
Gomez-Betancur D, Vargas-Bermudez DS, Giraldo-Ramírez S, Jaime J, Ruiz-Saenz J (2023) Canine circovirus: an emerging or an endemic undiagnosed enteritis virus? Front Vet Sci 10:1150636
Hsu HS, Lin TH, Wu HY, Lin LS, Chung CS, Chiou MT, Lin CN (2016) High detection rate of dog circovirus in diarrheal dogs. BMC Vet Res 12:116
Kapoor A, Dubovi EJ, Henriquez-Rivera JA, Lipkin WI (2012) Complete genome sequence of the first canine circovirus. J Virol 86:7018
Li L, McGraw S, Zhu K, Leutenegger CM, Marks SL, Kubiski S, Gaffney P, Dela Cruz FN Jr, Wang C, Delwart E, Pesavento PA (2013) Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg Infect Dis 19:534–541
Piewbang C, Jo WK, Puff C, van der Vries E, Kesdangsakonwut S, Rungsipipat A, Kruppa J, Jung K, Baumgartner W, Techangamsuwan S, Ludlow M, Osterhaus A (2018) Novel canine circovirus strains from Thailand: evidence for genetic recombination. Sci Rep 8:7524
Anderson A, Hartmann K, Leutenegger CM, Proksch AL, Mueller RS, Unterer S (2017) Role of canine circovirus in dogs with acute haemorrhagic diarrhoea. Vet Rec 180:542
Finn DR, Maldonado J, de Martini F, Yu J, Penton CR, Fontenele RS, Schmidlin K, Kraberger S, Varsani A, Gile GH, Barker B, Kollath DR, Muenich RL, Herckes P, Fraser M, Garcia-Pichel F (2021) Agricultural practices drive biological loads, seasonal patterns and potential pathogens in the aerobiome of a mixed-land-use dryland. Sci Total Environ 798:149239
Cerna GM, Serieys LEK, Riley SPD, Richet C, Kraberger S, Varsani A (2023) A circovirus and cycloviruses identified in feces of bobcats (Lynx rufus) in California. Arch Virol 168:23
Wu Z, Lu L, Du J, Yang L, Ren X, Liu B, Jiang J, Yang J, Dong J, Sun L, Zhu Y, Li Y, Zheng D, Zhang C, Su H, Zheng Y, Zhou H, Zhu G, Li H, Chmura A, Yang F, Daszak P, Wang J, Liu Q, Jin Q (2018) Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 6:178
Legnardi M, Grassi L, Franzo G, Menandro ML, Tucciarone CM, Minichino A, Dipineto L, Borrelli L, Fioretti A, Cecchinato M (2022) Detection and molecular characterization of a novel species of circovirus in a tawny owl (Strix aluco) in Southern Italy. Animals (Basel) 12:135
Wu Z, Yang L, Ren X, He G, Zhang J, Yang J, Qian Z, Dong J, Sun L, Zhu Y, Du J, Yang F, Zhang S, Jin Q (2016) Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J 10:609–620
Payne N, Kraberger S, Fontenele RS, Schmidlin K, Bergeman MH, Cassaigne I, Culver M, Varsani A, Van Doorslaer K (2020) novel circoviruses detected in feces of sonoran felids. Viruses 12:1027
Tang HB, Chen F, Rao G, Bai A, Jiang J, Du Y, Ren P, Liu J, Qin S, Yang L, Wu J (2017) Characterization of Akabane virus from domestic bamboo rat, Southern China. Vet Microbiol 207:280–285
Phan TG, da Costa AC, Del Valle MJ, Bucardo-Rivera F, Nordgren J, O’Ryan M, Deng X, Delwart E (2016) The fecal virome of South and Central American children with diarrhea includes small circular DNA viral genomes of unknown origin. Arch Virol 161:959–966
Siqueira JD, Dominguez-Bello MG, Contreras M, Lander O, Caballero-Arias H, Xutao D, Noya-Alarcon O, Delwart E (2018) Complex virome in feces from Amerindian children in isolated Amazonian villages. Nat Commun 9:4270
Kapusinszky B, Ardeshir A, Mulvaney U, Deng X, Delwart E (2017) Case-control comparison of enteric viromes in captive rhesus macaques with acute or idiopathic chronic diarrhea. J Virol. https://doi.org/10.1128/JVI.00952-17
Pearson VM, Caudle SB, Rokyta DR (2016) Viral recombination blurs taxonomic lines: examination of single-stranded DNA viruses in a wastewater treatment plant. PeerJ 4:e2585
de la Higuera I, Kasun GW, Torrance EL, Pratt AA, Maluenda A, Colombet J, Bisseux M, Ravet V, Dayaram A, Stainton D, Kraberger S, Zawar-Reza P, Goldstien S, Briskie JV, White R, Taylor H, Gomez C, Ainley DG, Harding JS, Fontenele RS, Schreck J, Ribeiro SG, Oswald SA, Arnold JM, Enault F, Varsani A, Stedman KM (2020) unveiling crucivirus diversity by mining metagenomic data. MBio 11:10–1128
Dayaram A, Galatowitsch ML, Arguello-Astorga GR, van Bysterveldt K, Kraberger S, Stainton D, Harding JS, Roumagnac P, Martin DP, Lefeuvre P, Varsani A (2016) Diverse circular replication-associated protein encoding viruses circulating in invertebrates within a lake ecosystem. Infect Genet Evol 39:304–316
Zhao M, Yue C, Yang Z, Li Y, Zhang D, Zhang J, Yang S, Shen Q, Su X, Qi D, Ma R, Xiao Y, Hou R, Yan X, Li L, Zhou Y, Liu J, Wang X, Wu W, Zhang W, Shan T, Liu S (2022) Viral metagenomics unveiled extensive communications of viruses within giant pandas and their associated organisms in the same ecosystem. Sci Total Environ 820:153317
Wang Y, Yang S, Liu D, Zhou C, Li W, Lin Y, Wang X, Shen Q, Wang H, Li C, Zong M, Ding Y, Song Q, Deng X, Qi D, Zhang W, Delwart E (2019) The fecal virome of red-crowned cranes. Arch Virol 164:3–16
Zhao G, Vatanen T, Droit L, Park A, Kostic AD, Poon TW, Vlamakis H, Siljander H, Harkonen T, Hamalainen AM, Peet A, Tillmann V, Ilonen J, Wang D, Knip M, Xavier RJ, Virgin HW (2017) Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci USA 114:E6166–E6175
Orton JP, Morales M, Fontenele RS, Schmidlin K, Kraberger S, Leavitt DJ, Webster TH, Wilson MA, Kusumi K, Dolby GA, Varsani A (2020) Virus discovery in desert tortoise fecal samples: novel circular single-stranded DNA viruses. Viruses 12:13
Palmieri N, de Jesus RM, Hess M, Bilic I (2021) Complete genomes of the eukaryotic poultry parasite Histomonas meleagridis: linking sequence analysis with virulence/attenuation. BMC Genom 22:753
Beer LC, Petrone-Garcia VM, Graham BD, Hargis BM, Tellez-Isaias G, Vuong CN (2022) Histomonosis in poultry: a comprehensive review. Front Vet Sci 9:880738
Dayaram A, Potter KA, Pailes R, Marinov M, Rosenstein DD, Varsani A (2015) Identification of diverse circular single-stranded DNA viruses in adult dragonflies and damselflies (Insecta: Odonata) of Arizona and Oklahoma, USA. Infect Genet Evol 30:278–287
Tochetto C, Muterle Varela AP, Alves de Lima D, Loiko MR, Mengue Scheffer C, Pinto Paim W, Cerva C, Schmidt C, Cibulski SP, Cano Ortiz L, Callegari Jacques SM, Franco AC, Quoos Mayer F, Roehe PM (2020) Viral DNA genomes in sera of farrowing sows with or without stillbirths. PLoS ONE 15:e0230714
Kinsella CM, Deijs M, Gittelbauer HM, van der Hoek L, van Dijk K (2022) Human clinical isolates of pathogenic fungi are host to diverse mycoviruses. Microbiol Spectr 10:e0161022
Karnkowska A, Vacek V, Zubacova Z, Treitli SC, Petrzelkova R, Eme L, Novak L, Zarsky V, Barlow LD, Herman EK, Soukal P, Hroudova M, Dolezal P, Stairs CW, Roger AJ, Elias M, Dacks JB, Vlcek C, Hampl V (2016) A eukaryote without a mitochondrial organelle. Curr Biol 26:1274–1284
Varsani A, Opriessnig T, Celer V, Maggi F, Okamoto H, Blomstrom AL, Cadar D, Harrach B, Biagini P, Kraberger S (2021) Taxonomic update for mammalian anelloviruses (family Anelloviridae). Arch Virol 166:2943–2953
Biagini P (2009) Classification of TTV and related viruses (anelloviruses). Curr Top Microbiol Immunol 331:21–33
Biagini P, Bendinelli M, Hino S, Kakkola L, Mankertz A, Niel C, Okamoto H, Raidal S, Teo CG, Todd D (2012) Family Anelloviridae. In: King AMQ, Adams EB, Carstens EB, E.J. L (eds) Virus taxonomy: ninth report of the International Committee on taxonomy of viruses. Academic press, London, pp 331–341
Butkovic A, Kraberger S, Smeele Z, Martin DP, Schmidlin K, Fontenele RS, Shero MR, Beltran RS, Kirkham AL, Aleamotu’a M, Burns JM, Koonin EV, Varsani A, Krupovic M (2023) Evolution of anelloviruses from a circovirus-like ancestor through gradual augmentation of the jelly-roll capsid protein. Virus Evol 9:vead035
Ng TF, Willner DL, Lim YW, Schmieder R, Chau B, Nilsson C, Anthony S, Ruan Y, Rohwer F, Breitbart M (2011) Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS ONE 6:e20579
Dennis TPW, de Souza WM, Marsile-Medun S, Singer JB, Wilson SJ, Gifford RJ (2019) The evolution, distribution and diversity of endogenous circoviral elements in vertebrate genomes. Virus Res 262:15–23
Liu H, Li P, Liu J (2011) Numerical investigation of underlying tunnel heave during a new tunnel construction. Tunn Undergr Space Technol 26:276–283
Patterson QM, Kraberger S, Martin DP, Shero MR, Beltran RS, Kirkham AL, Aleamotu’a M, Ainley DG, Kim S, Burns JM, Varsani A (2021) Circoviruses and cycloviruses identified in Weddell seal fecal samples from McMurdo Sound, Antarctica. Infect Genet Evol 95:105070
Dalton CS, van de Rakt K, Fahlman A, Ruckstuhl K, Neuhaus P, Popko R, Kutz S, van der Meer F (2017) Discovery of herpesviruses in Canadian wildlife. Arch Virol 162:449–456
Giacinti JA, Pearl DL, Ojkic D, Campbell GD, Jardine CM (2022) Genetic characterization of canine distemper virus from wild and domestic animal submissions to diagnostic facilities in Canada. Prev Vet Med 198:105535
Lopez-Perez AM, Moreno K, Chaves A, Ibarra-Cerdena CN, Rubio A, Foley J, List R, Suzan G, Sarmiento RE (2019) Carnivore protoparvovirus 1 at the wild-domestic carnivore interface in Northwestern Mexico. EcoHealth 16:502–511
Condori RE, Aragon A, Breckenridge M, Pesko K, Mower K, Ettestad P, Melman S, Velasco-Villa A, Orciari LA, Yager P, Streicker DG, Gigante CM, Morgan C, Wallace R, Li Y (2022) Divergent rabies virus variant of probable bat origin in 2 gray foxes, New Mexico, USA. Emerg Infect Dis 28:1137–1145
Pepin KM, Davis AJ, Streicker DG, Fischer JW, VerCauteren KC, Gilbert AT (2017) Predicting spatial spread of rabies in skunk populations using surveillance data reported by the public. PLoS Negl Trop Dis 11:e0005822
Wadhwa A, Wilkins K, Gao J, Condori Condori RE, Gigante CM, Zhao H, Ma X, Ellison JA, Greenberg L, Velasco-Villa A, Orciari L, Li Y (2017) A pan-lyssavirus taqman real-time RT-PCR assay for the detection of highly variable rabies virus and other lyssaviruses. PLoS Negl Trop Dis 11:e0005258
Kuzmin IV, Shi M, Orciari LA, Yager PA, Velasco-Villa A, Kuzmina NA, Streicker DG, Bergman DL, Rupprecht CE (2012) Molecular inferences suggest multiple host shifts of rabies viruses from bats to mesocarnivores in Arizona during 2001–2009. PLoS Pathog 8:e1002786
Velasco-Villa A, Reeder SA, Orciari LA, Yager PA, Franka R, Blanton JD, Zuckero L, Hunt P, Oertli EH, Robinson LE, Rupprecht CE (2008) Enzootic rabies elimination from dogs and reemergence in wild terrestrial carnivores, United States. Emerg Infect Dis 14:1849–1854
Real LA, Henderson JC, Biek R, Snaman J, Jack TL, Childs JE, Stahl E, Waller L, Tinline R, Nadin-Davis S (2005) Unifying the spatial population dynamics and molecular evolution of epidemic rabies virus. Proc Natl Acad Sci USA 102:12107–12111
Nadin-Davis SA, Abdel-Malik M, Armstrong J, Wandeler AI (2002) Lyssavirus P gene characterisation provides insights into the phylogeny of the genus and identifies structural similarities and diversity within the encoded phosphoprotein. Virology 298:286–305
Kuzmina NA, Kuzmin I, Ellison J, Rupprecht CE (2013) Conservation of binding epitopes for monoclonal antibodies on the rabies virus glycoprotein. J Antivir Antiretrovir 5:37–43
Acknowledgements
We thank Angela Oreshkova and Zach Ziebarth for their assistance in collecting the scat that was used in this research. We would like to also acknowledge the support provided by the Arizona State University Natural History Collection and the Central Arizona Phoenix Long Term Ecological Research Project (grant number DEB-1832016) through the equipment and space used to enable this research. Savage Cree Hess was supported by an Arizona State University Graduate Fellowship Grant. We thank Dakota M. Rowsey (Arizona State University, USA) for the constructive feedback on the manuscript and equipment provided to process the scat for dietary analysis. We acknowledge that this research has taken place on or near the ancestral lands of 22 tribes within Arizona, such as Akimel O’odham.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Research involving human participants and/or animals
The research did not involve human participants or animals. This study is a non-invasive study sampling feces of coyotes.
Additional information
Handling Editor: Sheela Ramamoorthy.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hess, S.C., Weiss, K.C.B., Custer, J.M. et al. Identification of small circular DNA viruses in coyote fecal samples from Arizona (USA). Arch Virol 169, 12 (2024). https://doi.org/10.1007/s00705-023-05937-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05937-w