Abstract
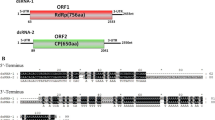
Rhizoctonia solani is a widely disseminated phytopathogen that is found in the soil and is capable of harming many important species of crops. Here, analysis of the R. solani AG-4 HG III strain A14 led to the identification of a novel mycovirus assigned the tentative name "Rhizoctonia solani partitivirus A14" (RsPV-A14), which was subjected to sequencing and associated analyses. This approach revealed that RsPV-A14 harbored two dsRNA segments, 2022 bp (dsRNA1) and 1905 bp (dsRNA2) in length. dsRNA1 was found to contain a single open reading frame (ORF1) that codes for a 622-amino-acid protein with conserved RNA-dependent RNA polymerase (RdRp) motifs, and dsRNA2 was found to contain an ORF (ORF2) that is predicted to code for a 558-amino-acid capsid protein (CP). BLASTp analysis using the putative RdRp of RsPV-A14 showed sequence similarity to partitiviruses, including Rosellinia necatrix partitivirus 7 (50.53% identity), an unclassified partitivirus. Phylogenetic analysis based on RdRp protein sequences suggested that RsPV-A14 is a novel member of the family Partitiviridae.


Similar content being viewed by others
Data availability
All relevant data are within the manuscript and its additional files.
References
González García V, Onco MAP, Susan VR (2006) Review. Biology and systematics of the form genus Rhizoctonia. Span J Agr Res 4:55–59
Yang YG, Zhao C, Guo ZJ, Wu XH (2015) Anastomosis group and pathogenicity of Rhizoctonia solani associated with stem canker and black scurf of potato in China. Eur J Plant Pathol 143:99–111
Sharon M, Kuninaga S, Hyakumachi M, Sneh B (2006) The advancing identification and classification of Rhizoctonia spp. using molecular and biotechnological methods compared with the classical anastomosis grouping. Mycoscience 47(6):299–316
Veldre V, Abarenkov K, Bahram M, Martos F, Selosse MA, Tamm H, Kõljalg U, Tedersoo L (2013) Evolution of nutritional modes of Ceratobasidiaceae (Cantharellales, Basidiomycota) as revealed from publicly available ITS sequences. Fungal Eco 6(4):256–268
Ogoshi A (1987) Ecology and pathogenicity of anastomosis and intraspecific groups of Rhizoctonia solani Kühn. Ann Rev Phytopathol 25:125–143
Gonzalez M, Pujol M, Metraux JP, Gonzalez-Garcia V, Borrás-Hidalgo O (2011) Tobacco leaf spot and root rot caused by Rhizoctonia solani kühn. Mol Plant Pathol 12:209–216
Razali NM, Hisham SN, Kumar IS, Shukla RN, Lee M, Abu Bakar MF, Nadarajah K (2021) Comparative genomics: insights on the pathogenicity and lifestyle of Rhizoctonia solani. Int J Mol Sci 22:2183
Marzano SL, Nelson BD, Ajayi-Oyetunde O, Bradley CA, Hughes TJ, Hartman GL, Eastburn DM, Domier LL (2016) Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J Virol 90:6846–6863
Abdoulaye AH, Foda MF, Kotta-Loizou I (2019) Viruses infecting the plant pathogenic fungus Rhizoctonia solani. Viruses. https://doi.org/10.3390/v11121113
Chen X, Yu Z, Sun Y, Yang M, Jiang N (2022) Molecular characterization of a novel partitivirus isolated from Rhizoctonia solani. Front Microbiol 13:978075
Vainio EJ, Chiba S, Ghabrial SA, Maiss E, Roossinck M, Sabanadzovic S, Suzuki N, Xie J, Nibert M (2018) ICTV Report Consortium. ICTV virus taxonomy profile: partitiviridae. J Gen Virol 99(1):17–18
Chen L, Chen JS, Zhang H, Chen SN (2006) Complete nucleotide sequences of three dsRNA segments from Raphanus sativus-root cv. Yidianhong with leaf yellow edge symptoms. Arch Virol 151(10):2077–2083
Liu W, Duns G, Chen J (2008) Genomic characterization of a novel partitivirus infecting Aspergillus ochraceus. Virus Genes 37(3):322–327
Tuomivirta TT, Hantula J (2003) Two unrelated double-stranded RNA molecule patterns in Gremmeniella abietina type A code for putative viruses of the families Totiviridae and Partitiviridae. Arch Virol 148(12):2293–2305
Nerva L, Ciuffo M, Vallino M, Margaria P, Varese GC, Gnavi G, Turina M (2016) Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res 219:22–38
Morris T, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Maan S, Rao S, Maan NS, Anthony SJ, Attoui H, Samuel AR, Mertens PP (2007) Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J Virol Methods 143:132–139
Li Y, Sun Y, Yu L, Chen W, Liu H, Yin L, Guang Y, Yang G, Mo X (2022) Complete genome sequence of a novel mitovirus from binucleate Rhizoctonia AG-K strain FAS2909W. Arch Virol 167:271–276
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027
Acknowledgements
This research was supported by the National Natural Scientific Foundation of China (32260036, 42067009), the Scientific Research Fund of the Education Department of Yunnan Province (2022Y696), and the Innovative Entrepreneurship Training Program for College Students (202205410226).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Handling Editor: Ioly Kotta-Loizou.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, R., Dong, B., Wang, Q. et al. Identification and nucleotide sequencing of a novel partitivirus derived from Rhizoctonia solani AG-4 HG III isolate A14. Arch Virol 168, 300 (2023). https://doi.org/10.1007/s00705-023-05923-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05923-2