Abstract
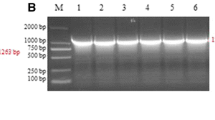
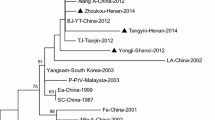
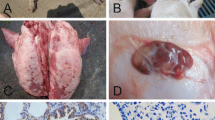
Pseudorabies virus (PRV) is an important pathogen that can cause harm to the pig population. Since 2011, there have been a number of large-scale outbreaks of pseudorabies on Chinese farms where animals had been vaccinated with the Bartha-K61 vaccine. In order to understand the epidemiological trend and genetic variations of PRV in Guangxi province, China, 819 tissue samples were collected from swine farms where PRV infection was suspected from 2013 to 2019, and these were tested for infectious wild strains of PRV. The results showed a positive rate of PRV in Guangxi province of 28.21% (231/819). Thirty-six wild-type PRV strains were successfully isolated from PRV-positive tissue samples, and a genetic evolutionary analysis was performed based on the gB, gC, gD, gE, and TK genes. Thirty of the PRV strains were found to be closely related to the Chinese variant strains HeN1-China-2012 and HLJ8-China-2013. In addition, five PRV strains were genetically related to Chinese classical strains, and one isolate was a recombinant of the PRV variant and the vaccine strain Bartha-K61. Amino acid sequence analysis showed that all 36 PRV strains had characteristic variant sites in the amino acid sequences of the gB, gC, gD, and gE proteins. Pathogenicity analysis showed that, compared to classical PRV strains, the PRV variant strains were more pathogenic in mice and had a lower LD50. Taken together, our results show that wild-type PRV infections are common on pig farms in Guangxi province of China and that the dominant prevalent strains were those of the PRV variants. The PRV variant strains also had increased pathogenicity in mice. Our data will provide a useful reference for understanding the prevalence and genetic evolution of PRV in China.




Similar content being viewed by others
References
Muller T, Hahn EC, Tottewitz F, Kramer M, Klupp BG, Mettenleiter TC, Freuling C (2011) Pseudorabies virus in wild swine: a global perspective. Arch Virol 156:1691–1705
Klupp BG, Hengartner CJ, Mettenleiter TC, Enquist LW (2004) Complete, annotated sequence of the pseudorabies virus genome. J Virol 78:424–440
Pomeranz LE, Reynolds AE, Hengartner CJ (2005) Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev 69:462–500
Laval K, Enquist LW (2020) The neuropathic itch caused by pseudorabies virus. Pathogens 9
Mettenleiter TC (2000) Aujeszky’s disease (pseudorabies) virus: the virus and molecular pathogenesis–state of the art, June 1999. Vet Res 31:99–115
Zheng HH, Fu PF, Chen HY, Wang ZY (2022) Pseudorabies virus: from pathogenesis to prevention strategies. Viruses 14
Yang H, Han H, Wang H, Cui Y, Liu H, Ding S (2019) A case of human viral encephalitis caused by pseudorabies virus infection in China. Front Neurol 10:534
Liu Q, Wang X, Xie C, Ding S, Yang H, Guo S, Li J, Qin L, Ban F, Wang D, Wang C, Feng L, Ma H, Wu B, Zhang L, Dong C, Xing L, Zhang J, Chen H, Yan R, Wang X, Li W (2021) A novel human acute encephalitis caused by pseudorabies virus variant strain. Clin Infect Dis 73:e3690–e3700
Freuling CM, Müller TF, Mettenleiter TC (2017) Vaccines against pseudorabies virus (PrV). Vet Microbiol 206:3–9
Zhou M, Wu X, Jiang D, Sui C, Chen L, Cong X, Xin X, Wang G, Li Y, Tian F, Chen Z, Zhang H, Qi J, Wang Z, Wu J, Shan H, Du Y (2019) Characterization of a moderately pathogenic pseudorabies virus variant isolated in China, 2014. Infect Genet Evol 68:161–171
Hu D, Zhang Z, Lv L, Xiao Y, Qu Y, Ma H, Niu Y, Wang G, Liu S (2015) Outbreak of variant pseudorabies virus in Bartha-K61-vaccinated piglets in central Shandong Province, China. J Vet Diagn Investig 27:600–605
Gu Z, Hou C, Sun H, Yang W, Dong J, Bai J, Jiang P (2015) Emergence of highly virulent pseudorabies virus in southern China. Can J Vet Res 79:221–228
An TQ, Peng JM, Tian ZJ, Zhao HY, Li N, Liu YM, Chen JZ, Leng CL, Sun Y, Chang D, Tong GZ (2013) Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg Infect Dis 19:1749–1755
Luo Y, Li N, Cong X, Wang CH, Du M, Li L, Zhao B, Yuan J, Liu DD, Li S, Li Y, Sun Y, Qiu HJ (2014) Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet Microbiol 174:107–115
Ishikawa K, Tsutsui M, Taguchi K, Saitoh A, Muramatsu M (1996) Sequence variation of the gC gene among pseudorabies virus strains. Vet Microbiol 49:267–272
Ye C, Zhang QZ, Tian ZJ, Zheng H, Zhao K, Liu F, Guo JC, Tong W, Jiang CG, Wang SJ, Shi M, Chang XB, Jiang YF, Peng JM, Zhou YJ, Tang YD, Sun MX, Cai XH, An TQ, Tong GZ (2015) Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: Evidence for the existence of two major genotypes. Virology 483:32–43
He W, Auclert LZ, Zhai X, Wong G, Zhang C, Zhu H, Xing G, Wang S, He W, Li K, Wang L, Han GZ, Veit M, Zhou J, Su S (2019) Interspecies transmission, genetic diversity, and evolutionary dynamics of pseudorabies virus. J Infect Dis 219:1705–1715
Bo Z, Miao Y, Xi R, Gao X, Miao D, Chen H, Jung YS, Qian Y, Dai J (2021) Emergence of a novel pathogenic recombinant virus from Bartha vaccine and variant pseudorabies virus in China. Transbound Emerg Dis 68:1454–1464
Sun Y, Luo Y, Wang CH, Yuan J, Li N, Song K, Qiu HJ (2016) Control of swine pseudorabies in China: opportunities and limitations. Vet Microbiol 183:119–124
Reed LJ, Muench H (1938) A simple method of estimating fifty per cent endpoints12. Am J Epidemiol 27:493–497
Ye C, Guo JC, Gao JC, Wang TY, Zhao K, Chang XB, Wang Q, Peng JM, Tian ZJ, Cai XH, Tong GZ, An TQ (2016) Genomic analyses reveal that partial sequence of an earlier pseudorabies virus in China is originated from a Bartha-vaccine-like strain. Virology 491:56–63
Huang J, Zhu L, Zhao J, Yin X, Feng Y, Wang X, Sun X, Zhou Y, Xu Z (2020) Genetic evolution analysis of novel recombinant pseudorabies virus strain in Sichuan, China. Transbound Emerg Dis 67:1428–1432
Fan J, Zeng X, Zhang G, Wu Q, Niu J, Sun B, Xie Q, Ma J (2016) Molecular characterization and phylogenetic analysis of pseudorabies virus variants isolated from Guangdong province of southern China during 2013–2014. J Vet Sci 17:369–375
Tan L, Yao J, Yang Y, Luo W, Yuan X, Yang L, Wang A (2021) Current status and challenge of pseudorabies virus infection in China. Virol Sin 36:588–607
Zhang C, Cui H, Zhang W, Meng L, Chen L, Wang Z, Zhao K, Chen Z, Qiao S, Liu J, Guo Z, Dong S (2022) Epidemiological investigation of porcine pseudorabies virus in Hebei Province, China, 2017–2018. Front Vet Sci 9:930871
Lin Y, Tan L, Wang C, He S, Fang L, Wang Z, Zhong Y, Zhang K, Liu D, Yang Q, Wang A (2021) Serological investigation and genetic characteristics of pseudorabies virus in Hunan Province of China From 2016 to 2020. Front Vet Sci 8:762326
Yao J, Li J, Gao L, He Y, Xie J, Zhu P, Zhang Y, Zhang X, Duan L, Yang S, Song C, Shu X (2022) Epidemiological investigation and genetic analysis of pseudorabies virus in Yunnan Province of China from 2017 to 2021. Viruses 14
Papageorgiou KV, Michailidou M, Grivas I, Petridou E, Stamelou E, Efraimidis K, Chen L, Drew TW, Kritas SK (2022) Bartha-K61 vaccine protects nursery pigs against challenge with novel european and asian strains of suid herpesvirus 1. Vet Res 53:47
Zhou J, Li S, Wang X, Zou M, Gao S (2017) Bartha-k61 vaccine protects growing pigs against challenge with an emerging variant pseudorabies virus. Vaccine 35:1161–1166
Wang J, Cui X, Wang X, Wang W, Gao S, Liu X, Kai Y, Chen C (2020) Efficacy of the Bartha-K61 vaccine and a gE(-)/gI(-)/TK(-) prototype vaccine against variant porcine pseudorabies virus (vPRV) in piglets with sublethal challenge of vPRV. Res Vet Sci 128:16–23
Ren Q, Li L, Pan H, Wang X, Gao Q, Huan C, Wang J, Zhang W, Jiang L, Gao S, Kai Y, Chen C (2022) Same dosages of rPRV/XJ5-gI(-)/gE(-)/TK(-) prototype vaccine or Bartha-K61 vaccine similarly protects growing pigs against lethal challenge of emerging vPRV/XJ-5 strain. Front Vet Sci 9:896689
Zhai X, Zhao W, Li K, Zhang C, Wang C, Su S, Zhou J, Lei J, Xing G, Sun H, Shi Z, Gu J (2019) Genome characteristics and evolution of pseudorabies virus strains in Eastern China from 2017 to 2019. Virol Sin 34:601–609
Sun Y, Liang W, Liu Q, Zhao T, Zhu H, Hua L, Peng Z, Tang X, Stratton CW, Zhou D, Tian Y, Chen H, Wu B (2018) Epidemiological and genetic characteristics of swine pseudorabies virus in mainland China between 2012 and 2017. PeerJ 6:e5785
Qin Y, Qin S, Huang X, Xu L, Ouyang K, Chen Y, Wei Z, Huang W (2023) Isolation and identification of two novel pseudorabies viruses with natural recombination or TK gene deletion in China. Vet Microbiol 280:109703
Acknowledgements
This research was funded by the National Natural Science Foundation (Grant no. 32260875). The authors would like to thank Dr. Dev Sooranna, Imperial College, London, for editing the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors report no conflict of interest.
Additional information
Handling Editor: Ana Cristina Bratanich.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, X., Qin, S., Wang, X. et al. Molecular epidemiological and genetic characterization of pseudorabies virus in Guangxi, China. Arch Virol 168, 285 (2023). https://doi.org/10.1007/s00705-023-05907-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05907-2