Abstract
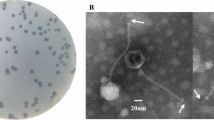
We isolated a K47-type Klebsiella pneumoniae phage from untreated hospital sewage: vB_KpnP_IME305 (GenBank no. OK149215). Next-generation sequencing (NGS) demonstrated that IME305 has a double-stranded DNA genome of 38,641 bp with 50.9% GC content. According to BLASTn comparisons, the IME305 genome sequence shares similarity with that of Klebsiella phage 6998 (97.37% identity and 95% coverage). IME305 contains 45 open reading frames (ORFs) and no rRNA, tRNA, or virulence-related gene sequences. Bioinformatic analysis showed that IME305 belongs to the phage subfamily Studiervirinae and genus Teetrevirus.


Similar content being viewed by others
References
(2022) Genomic analysis of Acinetobacter phage BUCT629, a newly isolated member of the genus Obolenskvirus. Arch Virol 167:1197–1199
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinf -OXFORD- 27:p1009–1010
Domingo-Calap P, Beamud B, Vienne J, González-Candelas F, Sanjuán R (2020) Isolation of Four Lytic Phages Infecting Klebsiella pneumoniae K22 Clinical Isolates from Spain. Multidisciplinary Digital Publishing Institute
Duffy C, Feiss M (2002) The large subunit of bacteriophage lambda’s terminase plays a role in DNA translocation and packaging termination. J Mol Biol 316:547–561
Gao M, Wang C, Qiang X, Liu H, Bai C (2020) Isolation and Characterization of a Novel Bacteriophage Infecting Carbapenem-Resistant Klebsiella pneumoniae. Curr Microbiol 77
Hatfull GF, Dedrick RM, Schooley RT (2022) Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu Rev Med 73:197–211
Jin X, Chen Q, Shen F, Jiang Y, Yu Y (2021) Resistance Evolution of Hypervirulent Carbapenem-resistant Klebsiella pneumoniae ST11 during Treatment with Tigecycline and Polymyxin. Emerg Microbes Infections :1–29
Khan Mirzaei M, Nilsson AS (2015) Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 10:e0118557
Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH (2016) Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front Microbiol 7:895
Lefkowitz EJ, Dempsey DM, Curtis HR, Orton RJ, Siddell SG, Smith DB (2018) Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Research:D708-D717
Liu Y, Leung S, Huang Y, Guo Y, Gao Z (2020) Identification of Two Depolymerases From Phage IME205 and Their Antivirulent Functions on K47 Capsule of Klebsiella pneumoniae. Frontiers in Microbiology 11
Liu Z, Gu Y, Li X, Liu Y, Ye Y, Guan S, Li J (2019) Identification and Characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Annals of laboratory medicine 39:167–175
Mi L, Liu Y, Wang C, He T, Gao S, Xing S, Huang Y, Fan H, Zhang X, Yu W (2019) Identification of a lytic Pseudomonas aeruginosa phage depolymerase and its anti-biofilm effect and bactericidal contribution to serum. Virus Genes
Mount DW (2007) Using the Basic Local Alignment Search Tool (BLAST). CSH protocols 2007:pdbtop17
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–214
Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y (2011) ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 66:307–312
Tomás M (2021) Phenotypic and Genomic Comparison of Klebsiella pneumoniae Lytic Phages: vB_KpnM-VAC66 and vB_KpnM-VAC13. Viruses 14
Wang C, Li P, Niu W, Yuan X, Liu H, Huang Y, An X, Fan H, Zhangxiang L, Mi L (2019) Protective and therapeutic application of the depolymerase derived from a novel KN1 genotype of Klebsiella pneumoniae bacteriophage in mice. Research in Microbiology
Wang C, Li P, Zhu Y, Huang Y, Bai C (2020) Identification of a Novel Acinetobacter baumannii Phage-Derived Depolymerase and Its Therapeutic Application in Mice. Frontiers in Microbiology 11
Wang Z, Cai R, Wang G, Guo Z, Liu X, Guan Y, Ji Y, Zhang H, Xi H, Zhao R (2021) Combination Therapy of Phage vB_KpnM_P-KP2 and Gentamicin Combats Acute Pneumonia Caused by K47 Serotype Klebsiella pneumoniae. Front Microbiol 12:674068
Zimmermann L, Stephens A, Nam SZ, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V (2018) A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol 430:2237–2243
Acknowledgements
This research was supported by the Anhui Natural Science Foundation Youth Program (2108085QH310), the Scientific Research Project of Anhui Health Committee (AHWJ2021b057), and the Research Fund Project of Anhui Medical University (2020xkj060).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All of the authors declare that they have no competing interests.
Nucleotide sequence accession number
The complete genome sequence of phage vB_KpnP_IME305 with annotations was submitted to the GenBank database under accession number OK149215.
Additional information
Communicated by Johannes Wittmann
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, C., Wang, Q., Mi, Z. et al. Genomic analysis of K47-type Klebsiella pneumoniae phage IME305, a newly isolated member of the genus Teetrevirus. Arch Virol 168, 280 (2023). https://doi.org/10.1007/s00705-023-05900-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05900-9