Abstract
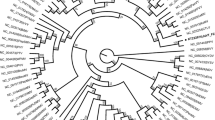
A new positive-sense, single-stranded RNA virus, tentatively named "Valeriana jatamansi tymovirus 1" (VaJV1, OQ730267), was isolated from Valeriana jatamansi Jones displaying symptoms of vein-clearing in Yunnan Province, China. The complete genome of VaJV1 consists of 6,215 nucleotides and contains three open reading frames (ORFs). The genome structure of VaJV1 is typical of members of the genus Tymovirus. BLASTn analysis and multiple sequence alignments showed that the complete genome and coat protein of VaJV1 shared the most sequence similarity (65.5% nucleotides and 50.5% amino acid sequence identity) with an isolate of the tymovirus okra mosaic virus (NC_009532). Phylogenetic analysis confirmed that VaJV1 clustered most closely with other tymoviruses. We propose that Valeriana jatamansi tymovirus 1 represents a new species within the genus Tymovirus.


Similar content being viewed by others
Data availability
The genomic sequence obtained in the current study is available in the GenBank database under accession number OQ730267.
References
Dhiman B, Sharma P, Pal PK (2020) Biology, chemical diversity, agronomy, conservation and industrial importance of Valeriana jatamansi: A natural sedative. J Appl Res Med Aromat Plants 16:100243
Wang C, Zheng Z, Deng X et al (2017) Flexible and powerful strategy for qualitative and quantitative analysis of valepotriates in Valeriana jatamansi Jones using high performance liquid chromatography with linear ion trap Orbitrap mass spectrometry. J Sep Sci 40:1906–1919
Sharma SAB, Kumari RAC, Kumar SAD et al (2016) Characterization of Cucumber mosaic virus isolates from Valeriana jatamansi, a medicinal herb in India(Note. J Plant Pathol 98(1):182
Zhe W, Xuhong S, Rex Frimpong A et al (2022) Detection of orthotospoviruses in medicinal plants in China. J Phytopathol 170(6):422–427
Sharma D, Sharma S, Singh N et al (2015) Valeriana jatamansi as a new natural host of Bhendi yellow vein mosaic virus and Papaya leaf curl virus betasatellite from Northern India. New Disease Reports 32:4
Hily JM, Candresse T, Garcia S et al (2018) High-throughput sequencing and the viromic study of grapevine leaves: from the detection of grapevine-infecting viruses to the description of a new environmental Tymovirales member. Front Microbiol 9:1782
Wang L, Lv X, Zhai Y et al (2012) Genomic characterization of a novel virus of the family Tymoviridae isolated from mosquitoes. PLoS ONE 7:e39845
Katsuma S, Tanaka S, Omuro N et al (2005) Novel macula-like virus identified in Bombyx mori cultured cells. J Virol 79:5577–5584
Martelli G, Sabanadzovic S, Abou Ghanem-Sabanadzovic N et al (2002) The family Tymoviridae. Adv Virol 147:1837–1846
Shouwei D, Jennifer H, Paul K et al (1990) The tymobox, a sequence shared by most tymoviruses: its use in molecular studies of tymoviruses. Nucleic Acids Res 18(5):1181–1187
Schirawski J, Voyatzakis A, Zaccomer B et al (2000) Identification and functional analysis of the turnip yellow mosaic tymovirus subgenomic promoter. J Virol 74:11073–11080
Shouwei D (1989) Molecular biology and evolution of tymoviruses, especially ononis yellow mosaic and kennedya yellow mosaic viruses. PhD thesis, The Australian National University, Canberra
Dreher TW (2004) Turnip yellow mosaic virus: transfer RNA mimicry, chloroplasts and a C-rich genome. Mol Plant Pathol 5(5):367–375
Dreher TW, Goodwin JB (1998) Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res 26(19):4356–4364
Bink HH, Schirawski J, Haenni AL et al (2003) The 5’-Proximal Hairpin of Turnip Yellow Mosaic Virus RNA: Its Role in Translation and Encapsidation. J Virol 77:7452–7458
Alabdullah A, Minafra A, Elbeaino T et al (2010) Complete nucleotide sequence and genome organization of Olive latent virus 3, a new putative member of the family Tymoviridae. Virus Res 152(1):10–18
Index of viruses—Tymoviridae (2009) ICTVdB—The Universal Virus Database. Columbia University, New York. http://www.ictvonline.org
Hayden CM, Mackenzie AM, Gibbs AJ (1998) Virion protein sequence variation among Australian isolates of turnip yellow mosaic tymovirus. Arch Virol 143:191–201
Stephan D, Siddiqua M, Ta Hoang A et al (2008) Complete nucleotide sequence and experimental host range of Okra mosaic virus. Virus Genes 36:231–240
Funding
No funding was received.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Ralf Georg Dietzgen
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chu, B., Anane, R.F., Li, S. et al. Complete genome sequence analysis of Valeriana jatamansi tymovirus 1: a novel member of the genus Tymovirus infecting Valeriana jatamansi Jones. Arch Virol 168, 245 (2023). https://doi.org/10.1007/s00705-023-05876-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05876-6