Abstract
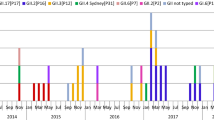
Noroviruses (NoVs) are a global concern, causing widespread outbreaks and sporadic acute gastroenteritis (AGE) cases across all age groups. Recent research has shed light on the emergence of novel recombinant strains of NoV in various countries. To delve deeper into this phenomenon, we extensively analyzed 1,175 stool samples collected from Japanese infants and children with AGE from six different prefectures in Japan over three years, from July 2018 to June 2021. Our investigation aimed to determine the prevalence and genetic characteristics of NoV associated with sporadic AGE while exploring the possibility of detecting NoV recombination events. Among the analyzed samples, we identified 355 cases positive for NoV, 11 cases attributed to GI genotypes, and 344 associated with GII genotypes. Notably, we discovered four distinct GI genotypes (GI.2, GI.3, GI.4, and GI.6) and seven diverse GII genotypes (GII.2, GII.3, GII.4, GII.6, GII.7, GII.14, and GII.17). The predominant genotypes were GII.4 (56.4%; 194 out of 344), followed by GII.2 and GII.3. Through dual genotyping based on sequencing of the ORF1/ORF2 junction region, we identified a total of 14 different RdRp/capsid genotypes. Of particular interest were the prevalent recombinant genotypes GII.4[P31] and GII.2[P16]. Notably, our study revealed a decrease in the number of children infected with NoV during and after the COVID-19 pandemic. These findings underscore the importance of continuous NoV surveillance efforts.






Similar content being viewed by others
Data Availability
A data availability statement has been already included in the Materials and Methods section (Nucleotide sequence accession numbers).
Abbreviations
- AGE:
-
Acute gastroenteritis
- COVID-19:
-
Coronavirus disease 2019
- ORFs:
-
Open reading frames
- RdRp:
-
RNA-dependent RNA polymerase
- VP:
-
Viral protein
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- bp:
-
Base pair
- BLAST:
-
Basic Local Alignment Search Tool
- RDP4:
-
Recombination Detection Program v.4.71
References
Afework DT, Shumie MK, Endalew GF, Adugna AG, Tarekegn BG (2022) Pooled prevalence and genetic diversity of norovirus in Africa: a systematic review and meta-analysis. Virol J 19:115. https://doi.org/10.1186/s12985-022-01835-w
Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA (2014) Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 14:725–730. https://doi.org/10.1016/S1473-3099(14)70767-4
Bon F, Ambert-Balay K, Giraudon H, Kaplon J, Le Guyader S, Pommepuy M, Gallay A, Vaillant V, de Valk H, Chikhi-Brachet R, Flahaut A, Pothier P, Kohli E (2005) Molecular epidemiology of caliciviruses detected in sporadic and outbreak cases of gastroenteritis in France from December 1998 to February 2004. J Clin Microbiol 43:4659–4664. https://doi.org/10.1128/JCM.43.9.4659-4664.2005
Bruggink LD (2022) Changes in norovirus incidence in Victoria, Australia, during the COVID-19 pandemic, 2020–2021. Commun Dis Intell. https://doi.org/10.33321/cdi.2022.46.61
Bull RA, Hansman GS, Clancy LE, Tanaka MM, Rawlinson WD, White PA (2005) Norovirus recombination in ORF1/ORF2 overlap. Emerg Infect Dis 11:1079–1085. https://doi.org/10.3201/eid1107.041273
Cannon JL, Bonifacio J, Bucardo F, Buesa J, Bruggink L, Chan MC, Fumian TM, Giri S, Gonzalez MD, Hewitt J, Lin JH, Mans J, Munoz C, Pan CY, Pang XL, Pietsch C, Rahman M, Sakon N, Selvarangan R, Browne H, Barclay L, Vinje J (2021) Global trends in norovirus genotype distribution among children with acute gastroenteritis. Emerg Infect Dis 27:1438–1445. https://doi.org/10.3201/eid2705.204756
Cao R, Ma X, Pan M (2022) Molecular characteristics of norovirus in sporadic and outbreak cases of acute gastroenteritis and in sewage in Sichuan, China. Virol J 19:180. https://doi.org/10.1186/s12985-022-01897-w
Chan-It W, Thongprachum A, Okitsu S, Nishimura S, Kikuta H, Baba T, Yamamoto A, Sugita K, Hashira S, Tajima T, Mizuguchi M, Ushijima H (2011) Detection and genetic characterization of norovirus infections in children with acute gastroenteritis in Japan, 2007–2009. Clin Lab 57:213–220
Chen C, Yan JB, Wang HL, Li P, Li KF, Wu B, Zhang H (2018) Molecular epidemiology and spatiotemporal dynamics of norovirus associated with sporadic acute gastroenteritis during 2013–2017, Zhoushan Islands, China. PLoS One 13:e0200911. https://doi.org/10.1371/journal.pone.0200911
Chhabra P, Browne H, Huynh T, Diez-Valcarce M, Barclay L, Kosek MN, Ahmed T, Lopez MR, Pan CY, Vinje J (2021) Single-step RT-PCR assay for dual genotyping of GI and GII norovirus strains. J Clin Virol 134:104689. https://doi.org/10.1016/j.jcv.2020.104689
Fukuda Y, Tsugawa T, Nagaoka Y, Ishii A, Nawa T, Togashi A, Kunizaki J, Hirakawa S, Iida J, Tanaka T, Kizawa T, Yamamoto D, Takeuchi R, Sakai Y, Kikuchi M, Nagai K, Asakura H, Tanaka R, Yoshida M, Hamada R, Kawasaki Y (2021) Surveillance in hospitalized children with infectious diseases in Japan: Pre- and post-coronavirus disease 2019. J Infect Chemother 27:1639–1647. https://doi.org/10.1016/j.jiac.2021.07.024
Honjo S, Kuronuma K, Fujiya Y, Nakae M, Ukae S, Nihira H, Yamamoto M, Akane Y, Kondo K, Takahashi S, Kimura H, Tsutsumi H, Kawasaki Y, Tsugawa T (2022) Genotypes and transmission routes of noroviruses causing sporadic acute gastroenteritis among adults and children, Japan, 2015–2019. Infect Genet Evol 104:105348. https://doi.org/10.1016/j.meegid.2022.105348
Imai N, Gaythorpe KAM, Bhatia S, Mangal TD, Cuomo-Dannenburg G, Unwin HJT, Jauneikaite E, Ferguson NM (2022) COVID-19 in Japan, January-March 2020: insights from the first three months of the epidemic. BMC Infect Dis 22:493. https://doi.org/10.1186/s12879-022-07469-1
Inasaki N, Aoyagi Y, Morioka S, Hasegawa S, Yoneda T, Saga Y, Itamochi M, Obuchi M (2019) A novel recombinant norovirus GII.4 Sydney 2012 strain detected from a food poisoning outbreak in the 2017–2018 season, Japan. Jpn J Infect Dis 72:64–67. https://doi.org/10.7883/yoken.JJID.2018.286
Jiang X, Wang M, Wang K, Estes MK (1993) Sequence and genomic organization of Norwalk virus. Virology 195:51–61. https://doi.org/10.1006/viro.1993.1345
Kanda N, Hashimoto H, Imai T, Yoshimoto H, Goda K, Mitsutake N, Hatakeyama S (2023) Indirect impact of the COVID-19 pandemic on the incidence of non-COVID-19 infectious diseases: a region-wide, patient-based database study in Japan. Public Health 214:20–24. https://doi.org/10.1016/j.puhe.2022.10.018
Khamrin P, Kumthip K, Supadej K, Thongprachum A, Okitsu S, Hayakawa S, Ushijima H, Maneekarn N (2017) Noroviruses and sapoviruses associated with acute gastroenteritis in pediatric patients in Thailand: increased detection of recombinant norovirus GII.P16/GII.13 strains. Arch Virol 162:3371–3380. https://doi.org/10.1007/s00705-017-3501-3
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lanata CF, Fischer-Walker CL, Olascoaga AC, Torres CX, Aryee MJ, Black RE (2013) Global causes of diarrheal disease mortality in children < 5 years of age: a systematic review. PLoS One 8:e72788. https://doi.org/10.1371/journal.pone.0072788
Lo M, Mitra S, De P, Banerjee A, Deb AK, Miyoshi SI, Manna A, Ghosh SK, Okamoto K, Dutta S, Chawla-Sarkar M (2021) Genetic characterization and evolutionary analysis of norovirus genotypes circulating among children in eastern India during 2018–2019. Arch Virol 166:2989–2998. https://doi.org/10.1007/s00705-021-05197-6
Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. https://doi.org/10.1128/JVI.73.1.152-160.1999
Mans J, Murray TY, Nadan S, Netshikweta R, Page NA, Taylor MB (2016) Norovirus diversity in children with gastroenteritis in South Africa from 2009 to 2013: GII.4 variants and recombinant strains predominate. Epidemiol Infect 144:907–916. https://doi.org/10.1017/S0950268815002150
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. https://doi.org/10.1093/ve/vev003
Medici MC, Tummolo F, Martella V, Giammanco GM, De Grazia S, Arcangeletti MC, De Conto F, Chezzi C, Calderaro A (2014) Novel recombinant GII.P16_GII.13 and GII.P16_GII.3 norovirus strains in Italy. Virus Res 188:142–145. https://doi.org/10.1016/j.virusres.2014.04.005
Nirwati H, Donato CM, Mawarti Y, Mulyani NS, Ikram A, Aman AT, Peppelenbosch MP, Soenarto Y, Pan Q, Hakim MS (2019) Norovirus and rotavirus infections in children less than five years of age hospitalized with acute gastroenteritis in Indonesia. Arch Virol 164:1515–1525. https://doi.org/10.1007/s00705-019-04215-y
Okitsu S, Hikita T, Thongprachum A, Khamrin P, Takanashi S, Hayakawa S, Maneekarn N, Ushijima H (2018) Detection and molecular characterization of two rare G8P[14] and G3P[3] rotavirus strains collected from children with acute gastroenteritis in Japan. Infect Genet Evol 62:95–108. https://doi.org/10.1016/j.meegid.2018.04.011
Pabbaraju K, Wong AA, Tipples GA, Pang XL (2019) Emergence of a Novel Recombinant Norovirus GII.P16-GII.12 Strain Causing Gastroenteritis, Alberta, Canada. Emerg Infect Dis 25:1556–1559. https://doi.org/10.3201/eid2508.190059
Pham NTK, Nishimura S, Shimizu-Onda Y, Trinh QD, Komine-Aizawa S, Khamrin P, Okitsu S, Sato S, Kobayashi T, Maneekarn N, Hayakawa S, Ushijima H (2022) Emerging norovirus GII.4 Sydney[P31] causing acute gastroenteritis outbreak in children in Japan, during COVID-19, 2021. J Infect Chemother 28:1347–1351. https://doi.org/10.1016/j.jiac.2022.05.015
Phengma P, Khamrin P, Jampanil N, Yodmeeklin A, Ushijima H, Maneekarn N, Kumthip K (2023) The emergence of recombinant norovirus GII.12[P16] and predominance of GII.3[P12] strains in pediatric patients with acute gastroenteritis in Thailand, 2019–2020. J Med Virol 95:e28321. https://doi.org/10.1002/jmv.28321
Portela AR, Hernandez JM, Bandeira RS, Junior ECS, de Melo TC, Lucena MSS, Teixeira DM, Siqueira JAM, Gabbay YB, Silva LD (2021) Retrospective molecular analysis of norovirus recombinant strains in the amazon region, Brazil. Infect Genet Evol 96:105130. https://doi.org/10.1016/j.meegid.2021.105130
Thongprachum A, Chan-it W, Khamrin P, Saparpakorn P, Okitsu S, Takanashi S, Mizuguchi M, Hayakawa S, Maneekarn N, Ushijima H (2014) Molecular epidemiology of norovirus associated with gastroenteritis and emergence of norovirus GII.4 variant 2012 in Japanese pediatric patients. Infect Genet Evol 23:65–73. https://doi.org/10.1016/j.meegid.2014.01.030
Thongprachum A, Okitsu S, Khamrin P, Maneekarn N, Hayakawa S, Ushijima H (2017) Emergence of norovirus GII.2 and its novel recombination during the gastroenteritis outbreak in Japanese children in mid-2016. Infect Genet Evol 51:86–88. https://doi.org/10.1016/j.meegid.2017.03.020
Utsumi T, Lusida MI, Dinana Z, Wahyuni RM, Soegijanto S, Soetjipto AAF, Sudarmo SM, Ranuh RG, Darma A, Juniastuti YLN, Doan YH, Shimizu H, Ishii K, Matsui C, Deng L, Abe T, Katayama K, Shoji I (2021) Molecular epidemiology and genetic diversity of norovirus infection in children hospitalized with acute gastroenteritis in East Java, Indonesia in 2015–2019. Infect Genet Evol 88:104703. https://doi.org/10.1016/j.meegid.2020.104703
van Beek J, de Graaf M, Al-Hello H, Allen DJ, Ambert-Balay K, Botteldoorn N, Brytting M, Buesa J, Cabrerizo M, Chan M, Cloak F, Di Bartolo I, Guix S, Hewitt J, Iritani N, Jin M, Johne R, Lederer I, Mans J, Martella V, Maunula L, McAllister G, Niendorf S, Niesters HG, Podkolzin AT, Poljsak-Prijatelj M, Rasmussen LD, Reuter G, Tuite G, Kroneman A, Vennema H, Koopmans MPG, NoroNet (2018) Molecular surveillance of norovirus, 2005–16: an epidemiological analysis of data collected from the NoroNet network. Lancet Infect Dis 18:545–553. https://doi.org/10.1016/S1473-3099(18)30059-8
Acknowledgements
We are grateful to Dr. Shuichi Nishimura, Dr. Hideaki Kikuta, Dr. Atsuko Yamamoto, Dr. Kumiko Sugita, Dr. Masaaki Kobayashi, and Dr. Tsuneyoshi Baba for collecting specimens, and we are also grateful to all parents and children who willingly participated in this study.
Funding
This study was supported by Grants-in-Aid for Japan Agency for Medical Research and Development (AMED) [grant number JP22fk0108122 and JP22wm0225006 to S.S., T.K., and H.U]; and the Nihon University Research Grant for 2022.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest is declared.
Additional information
Handling Editor: Akbar Dastjerdi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pham, N.T.K., Khamrin, P., Shimizu-Onda, Y. et al. Genetic diversity and declining norovirus prevalence in infants and children during Japan's COVID-19 pandemic: a three-year molecular surveillance. Arch Virol 168, 231 (2023). https://doi.org/10.1007/s00705-023-05856-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05856-w