Abstract
The spotted lanternfly (Lycorma delicatula) is an invasive pest that causes serious economic losses in fruit and wood production. Here, we identified a novel iflavirus named "Lycorma delicatula iflavirus 1" (LDIV1), in a spotted lanternfly. The full genome sequence of LDIV1 is 10,222 nt in length and encodes a polyprotein containing a picornavirus capsid-protein-domain-like domain, a cricket paralysis virus capsid superfamily domain, an RNA helicase domain, a peptidase C3 superfamily domain, and an RNA-dependent RNA polymerase (RdRp) domain. LDIV1 replicates in the host insect and activates small interfering RNA (siRNA)-based host antiviral immunity. Phylogenetic analysis demonstrated that LDIV1 is most closely related to an unspecified member of the order Picornavirales, with 61.7% sequence identity in the RdRp region and 57.6% sequence identity in the coat protein region, and thus meets the demarcation criteria for new species in the genus Iflavirus. To the best of our knowledge, LDIV1 is the first virus discovered in L. delicatula.


Similar content being viewed by others
Data availability
Raw reads generated by transcriptomic and sRNA sequencing have been deposited in the Genome Warehouse of the National Genomics Data Center (https://bigd.big.ac.cn/gwh) under accession number PRJCA009021.
References
Avanesyan A, Lamp WO (2020) Use of molecular gut content analysis to decipher the range of food plants of the invasive spotted lanternfly, Lycorma delicatula. Insects 11(4):215
Dara SK, Barringer L, Arthurs SP (2015) Lycorma delicatula (Hemiptera: Fulgoridae): a new invasive pest in the United States. J Integrated Pest Manag 6(1):20
Han JM, Kim H, Lim EJ, Lee S, Kwon YJ, Cho S (2008) Lycorma delicatula (Hemiptera: Auchenorrhyncha: Fulgoridae: Aphaeninae) finally, but suddenly arrived in Korea. Entomol Res 38(4):281–286
Kingan SB, Urban J, Lambert CC, Baybayan P, Childers AK, Coates B, Scheffler B, Hackett K, Korlach J, Geib SM (2019) A high-quality genome assembly from a single, field-collected spotted lanternfly (Lycorma delicatula) using the PacBio Sequel II system. GigaScience 8(10):122
van Oers MM (2010) Genomics and biology of Iflaviruses. Insect Vrol 231–250
Wu N, Zhang P, Liu W, Cao M, Wang X (2018) Sequence analysis and genomic organization of a new insect iflavirus, Sogatella furcifera honeydew virus 1. Adv Virol 163(7):2001–2003
Wang H, Liu Y, Liu W, Cao M, Wang X (2019) Full genome sequence of a novel iflavirus from the leafhopper Psammotettix alienus. Adv Virol 164(1):309–311
Ye ZX, Li YH, Chen JP, Zhang CX, Li JM (2021) Complete genome analysis of a novel iflavirus from a leaf beetle, Aulacophora lewisii. Arch Virol 166(1):309–312
Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, Mead A, Burroughs N, Evans DJ (2014) A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog 10(6):e1004230
Bailey L, Gibbs A, Woods R (1964) Sacbrood virus of the larval honey bee (Apis mellifera Linnaeus). Virology 23(3):425–429
Huang HJ, Ye ZX, Lu G, Zhang CX, Chen JP, Li JM (2021) Identification of salivary proteins in the whitefly Bemisia tabaci by transcriptomic and LC-MS/MS analyses. Insect Sci 28(5):1369–1381
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652
Sujeevan R, Hebert P, Diego F (2013) A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS ONE 8(7):e66213
Blum M, Chang H-Y, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi H, Natale DA, Necci M, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, Finn RD (2020) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49(D1):D344–D354
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359
Valles SM, Chen Y, Firth AE, Guerin DMA, Hashimoto Y, Herrero S, de Miranda JR, Ryabov E, Ictv Report C (2017) ICTV virus taxonomy profile: Iflaviridae. J Gen Virol 98(4):527–528
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35(21):4453–4455
Lu G, Ye ZX, He YJ, Zhang Y, Wang X, Huang HJ, Zhuo JC, Sun ZT, Yan F, Chen JP, Zhang CX, Li JM (2020) Discovery of two novel negeviruses in a dungfly collected from the Arctic. Viruses 12(7):692
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Nigg JC, Kuo YW, Falka BW (2020) Endogenous viral element-derived piwi-interacting RNAs (piRNAs) are not required for production of ping-pong-dependent piRNAs from Diaphorina citri densovirus. MBio 11(5):e02209-e2220
Zhang XD, Lu G, Ye ZX, Chen JP, Zhang CX, Li JM (2022) Complete genome analysis of a nege-like virus in aphids (Astegopteryx formosana). Adv Virol 167(1):267–270
Huang HJ, Ye ZX, Wang X, Yan XT, Zhang Y, He YJ, Qi YH, Zhang XD, Zhuo JC, Lu G, Lu JB, Mao QZ, Sun ZT, Yan F, Chen JP, Zhang CX, Li JM (2021) Diversity and infectivity of the RNA virome among different cryptic species of an agriculturally important insect vector: whitefly Bemisia tabaci. npj Biofilms Microbiomes 7(1):43
Funding
This research was supported by the National Natural Science Foundation of China (U20A2036) and the Ningbo Science and Technology Innovation 2025 Major Project (2019B10004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Simona Abba' .
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
705_2022_5503_MOESM1_ESM.tif
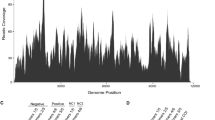
Assembly of RNA-seq reads to obtain the Lycorma delicatula iflavirus 1 (LDIV1) genome sequence. The RNA-seq dataset of L. delicatula (accession number: SRR5134712) was retrieved from NCBI SRA repository and mapped to the LDIV1 genome. (TIF 3480 KB)
Rights and permissions
About this article
Cite this article
Wang, YZ., Ye, ZX., Chen, J. et al. Complete genome analysis of a novel iflavirus from the spotted lanternfly Lycorma delicatula. Arch Virol 167, 2079–2083 (2022). https://doi.org/10.1007/s00705-022-05503-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05503-w