Abstract
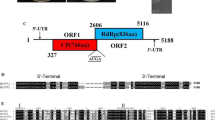
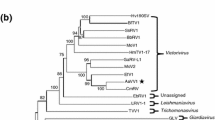
A novel victorivirus was detected in an isolate of Corynespora cassiicola strain 20180909-03 and was named "Corynespora cassiicola victorivirus 1" (CcVV1). The complete genome sequence of this virus is 5140 bp in length and contains 57% GC with two large open reading frames (ORFs) overlapping at the tetranucleotide AUGA. The ORFs were predicted to encode a coat protein (CP) and an RNA-dependent RNA polymerase (RdRp), respectively, which are conserved in dsRNA fungal viruses of the family Totiviridae. Comparison and phylogenetic analysis of the deduced amino acid sequences of RdRp and CP showed that CcVV1 is a new member of the genus Victorivirus. This is the first report of a genomic sequence of a victorivirus infecting Corynespora cassiicola.

Similar content being viewed by others
References
Smith LJOY (2008) Host range, phylogenetic, and pathogenic diversity of Corynespora cassiicola (Berk. & Curt.) Wei. Ph.D. dissertations, University of Florida, Gainesville, Florida, USA
Jia M, Liu X, Zhao H et al (2021) Cell-wall-degrading enzymes produced by sesame leaf spot pathogen Corynespora cassiicola. J Phytopathol 169:186–192. https://doi.org/10.1111/jph.12973
Shimomoto Y, Adachi R, Morita Y et al (2008) Corynespora blight of sweet pepper (Capsicum annuum) caused by Corynespora cassiicola (Berk. & Curt.) Wei. J Gen Plant Pathol 74:335–337. https://doi.org/10.1007/s10327-008-0100-x
Lakshmanan P, Jeyarajan R, Vidhyasekaran P (1990) A boll rot of cotton caused by Corynespora Cassiicola in Tamil Nadu, India. Phytoparasitica 18:171–173. https://doi.org/10.1007/BF02981234
Seaman WL, Shoemaker RA, Peterson EA (1965) PATHOGENICITY OF CORYNESPORA CASSIICOLA ON SOYBEAN. Can J Bot 43:1461–1469. https://doi.org/10.1139/b65-154
Sumabat LG, Kemerait RC, Brewer MT (2018) Phylogenetic Diversity and Host Specialization of Corynespora cassiicola Responsible for Emerging Target Spot Disease of Cotton and Other Crops in the Southeastern United States. Phytopathology® 108:892–901. https://doi.org/10.1094/PHYTO-12-17-0407-R
Yan XX, Yu CP, Fu XA et al (2016) CARD9 mutation linked to Corynespora cassiicola infection in a Chinese patient. Br J Dermatol 174:176–179. https://doi.org/10.1111/bjd.14082
Sato Y, Castón JR, Suzuki N (2018) The biological attributes, genome architecture and packaging of diverse multi-component fungal viruses. Curr Opin Virol 33:55–65. https://doi.org/10.1016/j.coviro.2018.07.009
Ghabrial SA, Castón JR, Jiang D et al (2015) 50-plus years of fungal viruses. Virology 479–480:356–368. https://doi.org/10.1016/j.virol.2015.02.034
Ghabrial SA, Nibert ML (2009) Victorivirus, a new genus of fungal viruses in the family Totiviridae. Arch Virol 154:373–379. https://doi.org/10.1007/s00705-008-0272-x
Jiang Y, Zhang T, Luo C et al (2015) Prevalence and diversity of mycoviruses infecting the plant pathogen Ustilaginoidea virens. Virus Res 195:47–56. https://doi.org/10.1016/j.virusres.2014.08.022
Liu H, Liu R, Li CX et al (2019) A victorivirus and two novel mitoviruses co-infected the plant pathogen nigrospora oryzae. Viruses 11. https://doi.org/10.3390/v11010083
Li W, Xia Y, Zhang H et al (2019) A Victorivirus from Fusarium asiaticum, the pathogen of Fusarium head blight in China. Arch Virol 164:313–316. https://doi.org/10.1007/s00705-018-4038-9
Kozlakidis Z, Herrero N, Coutts RHA (2013) The complete nucleotide sequence of a totivirus from Aspergillus foetidus. Arch Virol 158:263–266. https://doi.org/10.1007/s00705-012-1368-x
Wang J, Xiao Y, Liu X et al (2020) Complete genome sequence of a novel victorivirus isolated from the sesame charcoal rot fungus Macrophomina phaseolina. Arch Virol 165:509–514. https://doi.org/10.1007/s00705-019-04497-2
Sotaro C, Yu-Hsin L, Hideki K et al (2013) A Novel Victorivirus from a Phytopathogenic Fungus, Rosellinia necatrix, Is Infectious as Particles and Targeted by RNA Silencing. J Virol 87:6727–6738. https://doi.org/10.1128/JVI.00557-13
Huang S, Ghabrial SA (1996) Organization and expression of the double-stranded RNA genome of Helminthosporium victoriae 190S virus, a totivirus infecting a plant pathogenic filamentous fungus. Proc Natl Acad Sci U S A 93:12541–12546. https://doi.org/10.1073/pnas.93.22.12541
Li H, Havens WM, Nibert ML, Ghabrial SA (2015) An RNA cassette from Helminthosporium victoriae virus 190S necessary and sufficient for stop/restart translation. Virology 474:131–143. https://doi.org/10.1016/j.virol.2014.10.022
Wang Y, Zhao H, Xue C et al (2020) Complete genome sequence of a novel mycovirus isolated from the phytopathogenic fungus Corynespora cassiicola in China. Arch Virol 165:2401–2404. https://doi.org/10.1007/s00705-020-04723-2
Dodds JA (1983) Double-Stranded RNA from Plants Infected with Closteroviruses. Phytopathology 73:419. https://doi.org/10.1094/phyto-73-419
Lu S, Wang J, Chitsaz F et al (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268. https://doi.org/10.1093/nar/gkz991
Madeira F, Park Y, Lee J et al (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. https://doi.org/10.1093/nar/gkz268
Tamura K, Peterson D, Peterson N et al (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Kibbe WA (2007) OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35:W43–W46. https://doi.org/10.1093/nar/gkm234
Gasteiger E, Hoogland C, Gattiker A et al (2005) Protein Identification and Analysis Tools on the ExPASy Server. Proteom Protoc Handb 571–608. https://doi.org/10.1385/1592598900
Duvaud S, Gabella C, Lisacek F et al (2021) Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res 49:W216–W227. https://doi.org/10.1093/nar/gkab225
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. https://doi.org/10.1093/nar/gkg595
Liu W, Xie Y, Ma J et al (2015) IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31:3359–3361. https://doi.org/10.1093/bioinformatics/btv362
Sperschneider J, Datta A (2010) DotKnot: pseudoknot prediction using the probability dot plot under a refined energy model. Nucleic Acids Res 38:e103–e103. https://doi.org/10.1093/nar/gkq021
Wickner RB, Ghabrial SA, Nibert ML, Patterson JL, Wang CC (2011) Family Totiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus Taxonomy, 9th ICTV Report. Elsevier Inc, Amsterdam, pp 639–650
Acknowledgments
We are extremely grateful to Dr. Jing Wang for her valuable suggestions and help with sequence analysis.
Funding
This work was supported by the China Agriculture Research System of MOF and MARA [grant number CARS-14] and the Key Project of Science and Technology of Henan Province [grant number 201300110600].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Robert H.A. Coutts
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Rights and permissions
About this article
Cite this article
Liu, M., Ni, Y., Zhao, H. et al. Molecular characterization of a novel victorivirus infecting Corynespora cassiicola. Arch Virol 167, 1365–1368 (2022). https://doi.org/10.1007/s00705-022-05394-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05394-x