Abstract
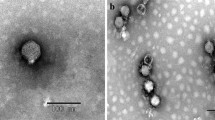
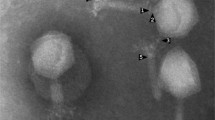
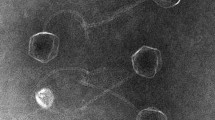
Salmonellosis is a disease of critical concern for public health, and the use of bacteriophages is among the most promising approaches to combating Salmonella. As Salmonella has various serotypes and strains, and bacteriophages are virulent to specific hosts, it is important to isolate phages and evaluate interactions with their hosts. In the present study, a novel Salmonella-infecting bacteriophage, pSal-SNUABM-01, was isolated and characterized. Transmission electron microscopy revealed that the bacteriophage is a member of the family Podoviridae and possesses an elongated head and a short tail. The phage genome is circular and 89,500 bp in size. A total of 162 open reading frames were predicted, eight of which were tRNAs. Morphological and genomic analysis revealed that pSal-SNUABM-01 is closely related to phage 7–11. In phylogenetic analysis, pSal-SNUABM-01 and 7–11 did not cluster together with the members of any established genus, suggesting that these two phages comprise a novel genus. The results of this study enhance our understanding of the phylogeny of the family Podoviridae and might be applicable to the development of bacteriophage treatments against Salmonella infections.


Similar content being viewed by others
References
Eng SK, Pusparajah P, Ab Mutalib NS, Ser HL, Chan KG, Lee LH (2015) Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci 8(3):284–293
Kwon J, Kim SG, Kim HJ, Giri SS, Kim SW, Lee SB, Park SC (2021) Bacteriophage as an alternative to prevent reptile-associated Salmonella transmission. Zoonoses Public Health 68(2):131–143
Feng P (1992) Commercial assay systems for detecting foodborne Salmonella: a review. J Food Prot 55(11):927–934
Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Frail A, Hoekstra RM, International Collaboration on Enteric Disease “Burden of Illness” Studies (2010) The global burden of nontyphoidal Salmonella gastroenteritis. Clin Inf Dis 50(6):882–889
Wessels K, Rip D, Gouws P (2021) Salmonella in chicken meat: consumption, outbreaks, characteristics, current control methods and the potential of bacteriophage use. Foods 10(8):1742
Aydin Demirarslan Ö, Alasalvar H, Yildirim Z (2021) Biocontrol of Salmonella Enteritidis on chicken meat and skin using lytic SE-P3, P16, P37, and P47 bacteriophages. LWT 137:110469
Shebs-Maurine EL, Giotto FM, Laidler ST, de Mello AS (2021) Effects of bacteriophages and peroxyacetic acid applications on beef contaminated with Salmonella during different grinding stages. Meat Science 173:108407
Xie Y, Thompson T, O’Leary C, Crosby S, Nguyen QX, Liu M, Gill JJ (2021) Differential bacteriophage efficacy in controlling Salmonella in cattle hide and soil models. Front Microbiol 12:1733
Oh JH, Park MK (2017) Recent trends in Salmonella outbreaks and emerging technology for biocontrol of Salmonella using phages in foods: a review. J Microbiol Biotechnol 27(12):2075–2088
Kim SG, Jun JW, Giri SS, Yun S, Kim HJ, Kim SW, Kang JW, Han SJ, Jeong DS, Park SC (2019) Isolation and characterisation of pVa-21, a giant bacteriophage with anti-biofilm potential against Vibrio alginolyticus. Sci Rep 9(1):1–10
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9(1):1–15
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29(12):2607–2618
Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V (2018) A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430(15):2237–2243
Lowe TM, Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44(W1):W54–W57
Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J (2009) DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25(1):119–120
Kropinski AM, Lingohr EJ, Ackermann HW (2011) The genome sequence of enterobacterial phage 7–11, which possesses an unusually elongated head. Adv Virol 156(1):149–151
Meier-Kolthoff JP, Göker M (2017) VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 33(21):3396–3404
Krumsiek J, Arnold R, Rattei T (2007) Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028
Myers EW, Miller W (1988) Optimal alignments in linear space. Bioinformatics 4(1):11–17
Turner D, Reynolds D, Seto D, Mahadevan P (2013) CoreGenes3.5: a webserver for the determination of core genes from sets of viral and small bacterial genomes. BMC Res Notes 6(1):1–4
Guzina J, Djordjevic M (2015) Inferring bacteriophage infection strategies from genome sequence: analysis of bacteriophage 7–11 and related phages. BMC Evol Biol 15(1):1–8
Acknowledgements
This research was supported by the Cooperative Research Program of the Center for Companion Animal Research (PJ0139852021) of the Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain studies with human participants or animals performed by any of the authors.
Additional information
Communicated by T. K. Frey.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kwon, J., Kim, S.G., Giri, S.S. et al. Genomic characterization of bacteriophage pSal-SNUABM-01, a novel elongated-head phage infecting Salmonella sp.. Arch Virol 167, 655–658 (2022). https://doi.org/10.1007/s00705-021-05342-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05342-1