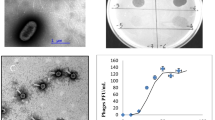
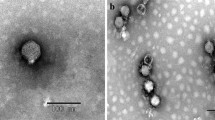
Abstract
Tailed bacteriophages have been at the center of attention, not only for their ability to infect and kill pathogenic bacteria but also due to their peculiar and intriguing complex contractile tail structure. Tailed bacteriophages with contractile tails are known to have a Myoviridae morphotype and are members of the order Caudovirales. Large bacteriophages with a genome larger than 150 kbp have been studied for their ability to use multiple infection and lysis strategies to replicate more efficiently. On the other hand, smaller bacteriophages with fewer genes are represented in the GenBank database in greater numbers, and have several genes with unknown function. Isolation and molecular characterization of a newly reported bacteriophage named Athena1 revealed that it is a strongly lytic bacteriophage with a genome size of 39,826 bp. This prompted us to perform a comparative genomic analysis of Vibrio myoviruses with a genome size of no more than 50 kbp. The results revealed a pattern of genomic organization that includes sets of genes responsible for virion morphogenesis, replication/recombination of DNA, and lysis/lysogeny switching. By studying phylogenetic gene markers, we were able to draw conclusions about evolutionary events that shaped the genomic mosaicism of these phages, pinpointing the importance of a conserved organization of the genomic region encoding the baseplate protein for successful infection of Gram-negative bacteria. In addition, we propose the creation of new genera for dwarf Vibrio myoviruses. Comparative genomics of phages infecting aquatic bacteria could provide information that is useful for combating fish pathogens in aquaculture, using novel strategies.










Similar content being viewed by others
Availability of data and material
The genome sequence of bacteriophage Athena1 is available in the GenBank database under the accession number MG640035. All raw data presented in the manuscript are available upon request.
References
Harada LK, Silva EC, Campos WF, Del Fiol FS, Vila M, Dąbrowska K, Krylov VN, Balcão VM (2018) Biotechnological applications of bacteriophages: state of the art. Microbiol Res 212:38–58
Zhou LQ, Bai JM (2010) Constructing genome phylogenetic tree of large dsDNA viruses using log-correlation distance. In: 2010 seventh international conference on fuzzy systems and knowledge discovery. IEEE, pp 2182–2185
Hendrix RW (2003) Bacteriophage genomics. Curr Opin Microbiol 6(5):506–511
Mavrich TN, Casey E, Oliveira J, Bottacini F, James K, Franz CM, Lugli GA, Neve H, Ventura M, Hatfull GF, Mahony J (2018) Characterization and induction of prophages in human gut-associated Bifidobacterium hosts. Sci Rep 8(1):1–17
Shapiro JW, Putonti C (2018) Gene co-occurrence networks reflect bacteriophage ecology and evolution. MBio 9(2):e01870-e1917
Ackermann HW (1998) Tailed bacteriophages: the order caudovirales. Adv Virus Res 51:135–201. https://doi.org/10.1016/s0065-3527(08)60785-x
Sullivan MB, Huang KH, Ignacio-Espinoza JC, Berlin AM, Kelly L, Weigele PR, DeFrancesco AS, Kern SE, Thompson LR, Young S, Yandava C (2010) Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol 12(11):3035–3056
Petrov VM, Nolan JM, Bertrand C, Levy D, Desplats C, Krisch HM, Karam JD (2006) Plasticity of the gene functions for DNA replication in the T4-like phages. J Mol Biol 361(1):46–68
Comeau AM, Tremblay D, Moineau S, Rattei T, Kushkina AI, Tovkach FI, Krisch HM, Ackermann HW (2012) Phage morphology recapitulates phylogeny: the comparative genomics of a new group of myoviruses. PloS One 7(7):e40102
Thompson JR, Polz MF (2006) Dynamics of Vibrio populations and their role in environmental nutrient cycling. In: The biology of Vibrios. American Society of Microbiology, pp 190–203
Letchumanan V, Chan KG, Pusparajah P, Saokaew S, Duangjai A, Goh BH, Ab Mutalib NS, Lee LH (2016) Insights into bacteriophage application in controlling Vibrio species. Front Microbiol 7:1114
Miller ES et al (2003) Complete genome sequence of the broad-host-range Vibrio phage KVP40: comparative genomics of a T4-related bacteriophage. J Bacteriol 185(17):5220–5233
Skliros D, Kalatzis PG, Katharios P, Flemetakis E (2016) Comparative functional genomic analysis of two Vibrio phages reveals complex metabolic interactions with the host cell. Front Microbiol 7:1807
Jang HB, Fagutao FF, Nho SW, Park SB, Cha IS, Yu JE, Lee JS, Im SP, Aoki T, Jung TS (2013) Phylogenomic network and comparative genomics reveal a diverged member of the ϕkz-related group, marine Vibrio phage ϕJM-2012. J Virol 87(23):12866–12878
Kalatzis PG, Rørbo NI, Castillo D, Mauritzen JJ, Jørgensen J, Kokkari C, Zhang F, Katharios P, Middelboe M (2017) Stumbling across the same phage: comparative genomics of widespread temperate phages infecting the fish pathogen Vibrio anguillarum. Viruses 9(5):122
Castillo D, D’Alvise P, Kalatzis PG, Kokkari C, Middelboe M, Gram L, Liu S, Katharios P (2015) Draft genome sequences of Vibrio alginolyticus strains V1 and V2, opportunistic marine pathogens. Genome Announc 3(4):e00729-e815
Kalatzis PG, Bastías R, Kokkari C, Katharios P (2016) Isolation and characterization of two lytic bacteriophages, φSt2 and φGrn1; phage therapy application for biological control of Vibrio alginolyticus in aquaculture live feeds. PLoS One 11(3):e0151101
Collins TJ (2007) ImageJ for microscopy. Biotechniques 43(S1):S25–S30
Shao Y, Wang IN (2008) Bacteriophage adsorption rate and optimal lysis time. Genetics 180(1):471–482
Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de 764 Bruijn graphs. Genome Res 18:821–829
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL (1999) Improved microbial gene 579 identification with GLIMMER. Nucleic Acids Res 27:4636–4641
Villa AA et al (2012) Complete genome sequence of Vibrio parahaemolyticus bacteriophage vB_VpaM_MAR
Oakey HJ, Cullen BR, Owens L (2002) The complete nucleotide sequence of the Vibrio harveyi bacteriophage VHML. J Appl Microbiol 93(6):1089–1098
Seguritan V, Feng IW, Rohwer F, Swift M, Segall AM (2003) Genome sequences of two closely related Vibrio parahaemolyticus phages, VP16T and VP16C. J Bacteriol 185(21):6434–6447
Leblanc C et al (2009) Isolation and genomic characterization of the first phage infecting Iodobacteria: ϕPLPE, a myovirus having a novel set of features. Environ Microbiol Rep 1(6):499–509
Popova AV, Zhilenkov EL, Myakinina VP, Krasilnikova VM, Volozhantsev NV (2012) Isolation and characterization of wide host range lytic bacteriophage AP22 infecting Acinetobacter baumannii. FEMS Microbiol Lett 332(1):40–46. https://doi.org/10.1111/j.1574-6968.2012.02573.x
Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J (2009) DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25(1):119–120
Krupovic M, Dutilh BE, Adriaenssens EM, Wittmann J, Vogensen FK, Sullivan MB, Rumnieks J, Prangishvili D, Lavigne R, Kropinski AM, Klumpp J (2016) Taxonomy of prokaryotic viruses: update from the ICTV bacterial and archaeal viruses subcommittee. Adv Virol 161(4):1095–1099
Zafar N, Mazumder R, Seto D (2002) CoreGenes: a computational tool for identifying and cataloging" core" genes in a set of small genomes. BMC Bioinform 3(1):1–8
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32(6):929–931
Veesler D, Cambillau C (2011) A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev 75(3):423–433
Darling AE, Tritt A, Eisen JA, Facciotti MT (2011) Mauve assembly metrics. Bioinformatics 27(19):2756–2757
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9(1):1–15
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42(D1):D206–D214
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S (2017) ViPTree: the viral proteomic tree server. Bioinformatics 33(15):2379–2380
Garneau JR, Depardieu F, Fortier LC, Bikard D, Monot M (2017) PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7(1):1–10
Wagemans J, Delattre AS, Uytterhoeven B, De Smet J, Cenens W, Aertsen A, Ceyssens PJ, Lavigne R (2015) Antibacterial phage ORFans of Pseudomonas aeruginosa phage LUZ24 reveal a novel MvaT inhibiting protein. Front Microbiol 6:1242
Niu YD, McAllister TA, Nash JH, Kropinski AM, Stanford K (2014) Four Escherichia coli O157: H7 phages: a new bacteriophage genus and taxonomic classification of T1-like phages. PloS One 9(6):e100426
Pride DT, Meinersmann RJ, Wassenaar TM, Blaser MJ (2003) Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res 13(2):145–158
Pride DT, Wassenaar TM, Ghose C, Blaser MJ (2006) Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genom 7(1):1–13
Casjens SR (2005) Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol 8(4):451–458
Rixon FJ, Schmid MF (2014) Structural similarities in DNA packaging and delivery apparatuses in Herpesvirus and dsDNA bacteriophages. Curr Opin Virol 5:105–110
Young ET, Menard RC, Harada JOHN (1981) Monocistronic and polycistronic bacteriophage T4 gene 23 messages. J Virol 40(3):790–799
Lin YR, Lin CS (2012) Genome-wide characterization of Vibrio phage ϕpp2 with unique arrangements of the mob-like genes. BMC Genom 13(1):224
Matamp N, Bhat SG (2019) Phage endolysins as potential antimicrobials against multidrug resistant Vibrio alginolyticus and Vibrio parahaemolyticus: current status of research and challenges ahead. Microorganisms. 7(3):84
Miller RV, Day M (2008) Contribution of lysogeny, pseudolysogeny, and starvation to phage ecology. Bacteriophage Ecol 114:143
López-Leal G, Reyes-Muñoz A, Santamaria RI, Cevallos MA, Pérez-Monter C, Castillo-Ramírez S (2021) A novel vieuvirus from multidrug-resistant Acinetobacter baumannii. Adv Virol 166(5):1401–1408
Keen EC (2015) A century of phage research: bacteriophages and the shaping of modern biology. BioEssays 37(1):6–9
Nobrega FL, Vlot M, de Jonge PA, Dreesens LL, Beaumont HJ, Lavigne R, Dutilh BE, Brouns SJ (2018) Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol 16(12):760–773
Cambillau C (2015) Bacteriophage module reshuffling results in adaptive host range as exemplified by the baseplate model of listerial phage A118. Virology 484:86–92
Krupovič M, Forterre P, Bamford DH (2010) Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J Mol Biol 397(1):144–160
Tarahovsky YS, Khusainov AA, Deev AA, Kim YV (1991) Membrane fusion during infection of Escherichia coli cells by phage T4. FEBS Lett 289(1):18–22
Han Y, Wang M, Liu Q et al (2019) Genome analysis of two novel lytic Vibrio maritimus phages isolated from the coastal surface seawater of Qingdao, China. Curr Microbiol 76:1225–1233. https://doi.org/10.1007/s00284-019-01736-2
Sternberg N, Coulby J (1990) Cleavage of the bacteriophage P1 packaging site (pac) is regulated by adenine methylation. Proc Natl Acad Sci 87(20):8070–8074
Bamford DH (2003) Do viruses form lineages across different domains of life? Res Microbiol 154(4):231–236
Cui J, Schlub TE, Holmes EC (2014) An allometric relationship between the genome length and virion volume of viruses. J Virol 88(11):6403–6410
Grayson P, Molineux IJ (2007) Is phage DNA ‘injected’ into cells—biologists and physicists can agree. Curr Opin Microbiol 10(4):401–409
Crummett LT, Puxty RJ, Weihe C, Marston MF, Martiny JB (2016) The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology 499:219–229
Konstantinidis KT, Tiedje JM (2004) Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci 101(9):3160–3165
Tang F, Bossers A, Harders F, Lu C, Smith H (2013) Comparative genomic analysis of twelve Streptococcus suis (pro) phages. Genomics 101(6):336–344
Perez Sepulveda B, Redgwell T, Rihtman B, Pitt F, Scanlan DJ, Millard A (2016) Marine phage genomics: the tip of the iceberg. FEMS Microbiol Lett 363(15):fnw158
Adriaenssens E, Brister JR (2017) How to name and classify your phage: an informal guide. Viruses 9(4):70
Turner D, Kropinski AM, Adriaenssens EM (2021) A roadmap for genome-based phage taxonomy. Viruses 13(3):506
Acknowledgments
We would like to thank Assistant Professor Gerasimos Daras for assisting with PhageTerm software analysis.
Funding
This research was co-financed by Greece and the European Union (European Social Fund- ESF) through the Operational Programme "Human Resources Development, Education and Lifelong Learning" in the context of the project “Reinforcement of Postdoctoral Researchers—2nd Cycle” (MIS-5033021), implemented by the State Scholarships Foundation (ΙΚΥ).
Author information
Authors and Affiliations
Contributions
DS and EF acquired funding. EF, DS and EK conceived the study. DS performed DNA extraction. DS and EK performed phage bioinformatic analysis and wrote the manuscript. PK assisted in TEM photography. PK and CK assisted in data curation. All authors critically reviewed the manuscript, commented on the final draft, and approved the version to be published.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest and no competing interests.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
705_2021_5340_MOESM3_ESM.docx
Supplementary Table S1 Coding DNA products of newly reported bacteriophage Athena1, including their protein_id and presence or absence of protein products in Vibrio phage VBM1 and reference phage PLPE (DOCX 16 KB)
Rights and permissions
About this article
Cite this article
Skliros, D., Karpouzis, E., Kalloniati, C. et al. Comparative genomic analysis of dwarf Vibrio myoviruses defines a conserved gene cluster for successful phage infection. Arch Virol 167, 501–516 (2022). https://doi.org/10.1007/s00705-021-05340-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05340-3