Abstract
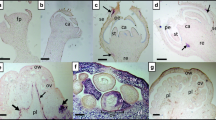
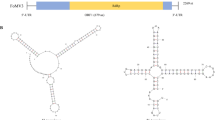
The complete genome sequences of two isolates of spiraea yellow leafspot virus (SYLSV) were determined. Spiraea (Spiraea x bumalda) ‘Anthony Waterer’ plants showing virus-like symptoms including yellow spotting and leaf deformation were used for sequencing. The viral genome of SYLSV-MN (Minnesota) and SYLSV-MD (Maryland) is 8,017bp in length. The sequences share 95% identity at the nucleotide level. Both isolates have the same genome organization containing three open reading frames (ORFs), with ORF3 being the largest, encoding a putative polyprotein of 232 kDa with conserved domains including a zinc finger, pepsin-like aspartate protease, reverse transcriptase (RT), and RNase H. Pairwise comparisons between members of the genus Badnavirus showed that gooseberry vein banding associated virus GB1 (HQ852248) and rubus yellow net virus isolate Baumforth's Seedling A (KM078034) were the closest related virus sequences to SYLSV, sharing 73% identity at the nucleotide level. Bacilliform virions with dimensions of 150 nm × 30 nm were observed in virus preparations from symptomatic, but not asymptomatic, plants.


Similar content being viewed by others
References
Lockhart BEL, Geering ADW (2002) Partial characterization of two aphid-transmitted viruses associated with yellow leafspot of Spiraea. Acta Hortic 568:163–168. https://doi.org/10.17660/ActaHortic.2002.568.24
Alvarez-Quinto RA, Lockhart BEL, Olszewski N (2019) Complete genome sequence of a previously undescribed badnavirus occurring in Polyscias fruticosa L. (Ming aralia). Arch Virol. https://doi.org/10.1007/s00705-019-04307-9
Teycheney PY, Geering ADW, Dasgupta I et al (2020) ICTV Virus taxonomy profile: Caulimoviridae. J Gen Virol 101:1025–1026
Geering ADW (2021) Badnaviruses (Caulimoviridae). In: Bamford DH, Zuckerman M (eds) Encyclopedia of virology. Academic P Elsevier, Oxford, pp 158–168
Funding
This work was supported by the USDA National Institute of Food and Agriculture, Hatch project #1009997 and the University of Minnesota, Twin Cities. Alvarez-Quinto R.A is supported by the Republic of Ecuador through the Ecuadorean Science and Technology Secretariat (SENESCYT).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All of the authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Elvira Fiallo-Olivé
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Alvarez-Quinto, R.A., Serrano, J., Olszewski, N. et al. Complete genome sequences of two isolates of spiraea yellow leafspot virus (genus Badnavirus) from Spiraea x bumalda ‘Anthony Waterer’. Arch Virol 167, 631–634 (2022). https://doi.org/10.1007/s00705-021-05336-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05336-z