Abstract
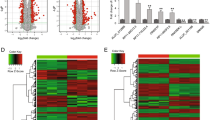
Coxsackievirus B3 (CVB3) is a major cause of viral myocarditis in humans. Although there have been studies on CVB3 infection and pathogenesis, the precise disease mechanism is still not clear. In this study, we used RNA-seq technology to compare the transcriptomic profile of virus-infected HeLa cells to that of uninfected cells to identify key genes involved in host-virus interaction. For this, two CVB3 strains, CVB3 Woodruff, an experimental strain, and GD16-69/GD/CHN/2016, a clinical strain, were selected to examine the common mechanisms underlying their infection. Transcriptomic profiles revealed increased expression of the cell cycle genes CCNG2, GADD45B, PIM1, RBM15, KLF10, and RIOK3 and decreased expression of CYBA. The autophagy-related genes ATG12 and YOD1 were found to be upregulated, while the expression of SOD2 and XPO1 increased slightly in infected cells, and only a minor change was observed in GABARAP expression. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed the FoxO signaling pathway to be enriched and showed a close interaction with differentially expressed genes (DEGs) in the protein-protein interaction network. DEGs associated with related pathways such as cell cycle, autophagy, and oxidative stress resistance were also confirmed by qRT-PCR. In summary, the FoxO signaling pathway was activated during infection with both CVB3 strains and was found to have a regulatory role in downstream pathways such as cell cycle, autophagy, oxidative stress resistance, and the antiviral immune response.





Similar content being viewed by others
References
Garmaroudi FS et al (2015) Coxsackievirus B3 replication and pathogenesis. Future Microbiol 10(4):629–653
Peischard S et al (2019) A kidnapping story: How coxsackievirus B3 and its host cell interact. Cell Physiol Biochem 53(1):121–140
Wang Y et al (2019) The capsid protein VP1 of coxsackievirus B induces cell cycle arrest by up-regulating heat shock protein 70. Front Microbiol 10:1633
Luo XN et al (2018) Coxsackievirus B3 infection triggers autophagy through 3 pathways of endoplasmic reticulum stress. Biomed Environ Sci 31(12):867–875
Knowlton KU et al (1996) A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J Virol 70(11):7811–7818
Lukashev AN (2005) Role of recombination in evolution of enteroviruses. Rev Med Virol 15(3):157–167
Han Z et al (2019) Two Coxsackievirus B3 outbreaks associated with hand, foot, and mouth disease in China and the evolutionary history worldwide. BMC Infect Dis 19(1):466
Huang H, Tindall DJ (2007) Dynamic FoxO transcription factors. J Cell Sci 120(Pt 15):2479–2487
Cheng Z (2019) The FoxO-autophagy axis in health and disease. Trends Endocrinol Metab 30(9):658–671
Williamson A et al (2009) Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci USA 106(43):18213–18218
Iadevaia V et al (2010) PIM1 kinase is destabilized by ribosomal stress causing inhibition of cell cycle progression. Oncogene 29(40):5490–5499
Rajamannan NM et al (2007) TGFbeta inducible early gene-1 (TIEG1) and cardiac hypertrophy: discovery and characterization of a novel signaling pathway. J Cell Biochem 100(2):315–325
Machitani M et al (2020) The RNA transport factor PHAX is required for proper histone H2AX expression and DNA damage response. RNA 26(11):1716–1725
Silla T et al (2020) The human ZC3H3 and RBM26/27 proteins are critical for PAXT-mediated nuclear RNA decay. Nucleic Acids Res 48(5):2518–2530
Tracy S, Gauntt C (2008) Group B coxsackievirus virulence. Curr Top Microbiol Immunol 323:49–63
Pons-Salort M, Parker EP, Grassly NC (2015) The epidemiology of non-polio enteroviruses: recent advances and outstanding questions. Curr Opin Infect Dis 28(5):479–487
Pan J et al (2011) Single amino acid changes in the virus capsid permit coxsackievirus B3 to bind decay-accelerating factor. J Virol 85(14):7436–7443
Schmidtke M et al (2000) Attachment of coxsackievirus B3 variants to various cell lines: mapping of phenotypic differences to capsid protein VP1. Virology 275(1):77–88
Selinka HC et al (2002) Comparative analysis of two coxsackievirus B3 strains: putative influence of virus-receptor interactions on pathogenesis. J Med Virol 67(2):224–233
Martino TA et al (1998) Cardiovirulent coxsackieviruses and the decay-accelerating factor (CD55) receptor. Virology 244(2):302–314
Jensen KJ et al (2013) An ERK-p38 subnetwork coordinates host cell apoptosis and necrosis during coxsackievirus B3 infection. Cell Host Microbe 13(1):67–76
Esfandiarei M et al (2007) Coxsackievirus B3 activates nuclear factor kappa B transcription factor via a phosphatidylinositol-3 kinase/protein kinase B-dependent pathway to improve host cell viability. Cell Microbiol 9(10):2358–2371
Accili D, Arden KC (2004) FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117(4):421–426
Ma Z et al (2020) Forkhead box O1-mediated ubiquitination suppresses RIG-I-mediated antiviral immune responses. Int Immunopharmacol 90:107152
Guo F et al (2018) Japanese encephalitis virus induces apoptosis by inhibiting FOXO signaling pathway. Vet Microbiol 220:73–82
León J et al (2014) Melatonin reduces endothelin-1 expression and secretion in colon cancer cells through the inactivation of FoxO-1 and NF-κβ. J Pineal Res 56(4):415–426
van Boxtel R et al (2013) FOXP1 acts through a negative feedback loop to suppress FOXO-induced apoptosis. Cell Death Differ 20(9):1219–1229
Mi L, Rong Bao G and Jun H (2021) Transcriptomic analysis of CVB3 induced acute viral myocarditis in C57BL/6 mouse model. Chin J Virol 37(3):12
Kim W et al (2011) PIM1-activated PRAS40 regulates radioresistance in non-small cell lung cancer cells through interplay with FOXO3a, 14-3-3 and protein phosphatases. Radiat Res 176(5):539–552
Yang Y et al (2012) Biological effects of decreasing RBM15 on chronic myelogenous leukemia cells. Leuk Lymphoma 53(11):2237–2244
Feng J et al (2014) RIOK3 is an adaptor protein required for IRF3-mediated antiviral type I interferon production. J Virol 88(14):7987–7997
Shi X et al (2016) Coxsackievirus B3 infection induces autophagic flux, and autophagosomes are critical for efficient viral replication. Arch Virol 161(8):2197–2205
Giansanti P et al (2020) Dynamic remodelling of the human host cell proteome and phosphoproteome upon enterovirus infection. Nat Commun 11(1):4332
Martin AP et al (2019) STK38 kinase acts as XPO1 gatekeeper regulating the nuclear export of autophagy proteins and other cargoes. EMBO Rep 20(11):e48150
Uddin MH, Zonder JA, Azmi AS (2020) Exportin 1 inhibition as antiviral therapy. Drug Discov Today 25:1775–1781
Papadopoulos C et al (2017) VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. Embo j 36(2):135–150
Zhang Z et al (2020) Acetylation-dependent deubiquitinase OTUD3 controls MAVS activation in innate antiviral immunity. Mol Cell 79(2):304–319 (e7)
Li Q et al (2014) Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog 10(5):e1004163
Ma N et al (2019) Oxidative stress-related gene polymorphisms are associated with hepatitis B virus-induced liver disease in the northern Chinese Han population. Front Genet 10:1290
Salmen S et al (2010) HIV-1 Nef associates with p22-phox, a component of the NADPH oxidase protein complex. Cell Immunol 263(2):166–171
Song QQ et al (2015) Coxsackievirus B3 2A protease promotes encephalomyocarditis virus replication. Virus Res 208:22–29
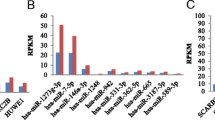
Yao HL et al (2019) Construction of miRNA-target networks using microRNA profiles of CVB3-infected HeLa cells. Sci Rep 9(1):17876
Kang H et al (2017) Vascular smooth muscle cell glycocalyx mediates shear stress-induced contractile responses via a Rho kinase (ROCK)-myosin light chain phosphatase (MLCP) pathway. Sci Rep 7:42092
Gao L et al (2019) Suppression of glioblastoma by a drug cocktail reprogramming tumor cells into neuronal like cells. Sci Rep 9(1):3462
Li J et al (2020) Comparative transcriptome analysis reveals different host cell responses to acute and persistent foot-and-mouth disease virus infection. Virol Sin 35(1):52–63
Chen H, Boutros PC (2011) VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform 12:35
Franceschini A et al (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41(Database issue):D808–D815
Young MD et al (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11(2):R14
Kanehisa M et al (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36(Database issue):D480–D484
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408
Acknowledgements
We appreciate the kind support from Wenbo Xu, Director of the National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, for the resources and information on CVB3 epidemic viral strains.
Funding
This article was not supported by any particular grant.
Author information
Authors and Affiliations
Contributions
M.L. and J.H. conceived and designed the experiments and revised the manuscript. M.L. performed the experiments, analyzed the data, and wrote the manuscript. Q.Y. contributed reagents and materials. As the corresponding author of this study, J.H. has access to all of the data from the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Handling Editor: Tim Skern.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, M., Yang, Q. & Han, J. Transcriptomic analysis reveals that coxsackievirus B3 Woodruff and GD strains use similar key genes to induce FoxO signaling pathway activation in HeLa cells. Arch Virol 167, 131–140 (2022). https://doi.org/10.1007/s00705-021-05292-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05292-8