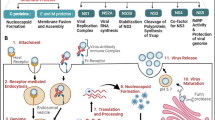
Abstract
Viruses cause a variety of diseases in humans and other organisms. The most important defense mechanism against viral infections is initiated when the viral genome is sensed by host proteins, and this results in interferon production and pro-inflammatory cytokine responses. The sensing of the viral genome or its replication intermediates within host cells is mediated by cytosolic proteins. For example, cGAS and IFI16 recognize non-self DNA, and RIG-I and MDA5 recognize non-self RNA. Once these sensors are activated, they trigger a cascade of reactions activating downstream molecules, which eventually results in the transcriptional activation of type I and III interferons, which play a critical role in suppressing viral propagation, either by directly limiting their replication or by inducing host cells to inhibit viral protein synthesis. The immune response against viruses relies solely upon sensing of viral genomes and their downstream signaling molecules. Although DNA and RNA viruses are sensed by distinct classes of receptor proteins, there is a possibility of overlap between the viral DNA and viral RNA sensing mechanisms. In this review, we focus on various host sensing molecules and discuss the associated signaling pathways that are activated in response to different viral infections. We further highlight the possibility of crosstalk between the cGAS-STING and the RIG-I-MAVS pathways to limit viral infections. This comprehensive review delineates the mechanisms by which different viruses evade host cellular responses to sustain within the host cells.



Similar content being viewed by others
References
Tan X, Sun L, Chen J, Chen ZJ (2018) Detection of microbial infections through innate immune sensing of nucleic acids. Annu Rev Microbiol 72:447–478
Roh JS, Sohn DH (2018) Damage-associated molecular patterns in inflammatory diseases. Immune Netw 18 (4)
Dempsey A, Bowie AG (2015) Innate immune recognition of DNA: a recent history. Virology 479:146–152
Mitchell G, Isberg RR (2017) Innate immunity to intracellular pathogens: balancing microbial elimination and inflammation. Cell Host Microbe 22(2):166–175
Ma Z, Damania B (2016) The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe 19(2):150–158
Ni G, Ma Z, Damania B (2018) cGAS and STING: at the intersection of DNA and RNA virus-sensing networks. PLoS Pathog 14(8):e1007148
Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M (2006) 5′-triphosphate RNA is the ligand for RIG-I. Science 314(5801):994–997
Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT (2004) TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol 5(2):190
Pasare C, Medzhitov R (2005) Toll-like receptors: linking innate and adaptive immunity. In: Mechanisms of lymphocyte activation and immune regulation X, pp 11–18
Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448(7152):501–505
Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C (2008) TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451(7179):725
Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu Y-J (2011) The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12(10):959
Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V (2009) RIG-I-dependent sensing of poly (dA: dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol 10(10):1065
Chiu Y-H, MacMillan JB, Chen ZJ (2009) RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138(3):576–591
Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11(11):997
Diner BA, Li T, Greco TM, Crow MS, Fuesler JA, Wang J, Cristea IM (2015) The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol Syst Biol 11(2):787
Crow MS, Cristea IM (2017) Human antiviral protein IFIX suppresses viral gene expression during herpes simplex virus 1 (HSV-1) infection and is counteracted by virus-induced proteasomal degradation. Mol Cell Proteomics 16(4 suppl 1):S200–S214
Sun L, Wu J, Du F, Chen X, Chen ZJ (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339(6121):786–791
Whiteley AT, Eaglesham JB, de Oliveira Mann CC, Morehouse BR, Lowey B, Nieminen EA, Danilchanka O, King DS, Lee AS, Mekalanos JJ (2019) Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567(7747):194–199
Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner K-P (2013) Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498(7454):332–337
Mankan AK, Schmidt T, Chauhan D, Goldeck M, Höning K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V (2014) Cytosolic RNA: DNA hybrids activate the cGAS–STING axis. EMBO J 33(24):2937–2946
Tao J, Zhang X-W, Jin J, Du X-X, Lian T, Yang J, Zhou X, Jiang Z, Su X-D (2017) Nonspecific DNA binding of cGAS N terminus promotes cGAS activation. J Immunol 198(9):3627–3636
Andreeva L, Hiller B, Kostrewa D, Lässig C, de Oliveira Mann CC, Drexler DJ, Maiser A, Gaidt M, Leonhardt H, Hornung V (2017) cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein–DNA ladders. Nature 549(7672):394
Herzner A-M, Hagmann CA, Goldeck M, Wolter S, Kübler K, Wittmann S, Gramberg T, Andreeva L, Hopfner K-P, Mertens C (2015) Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol 16(10):1025
Li X-D, Wu J, Gao D, Wang H, Sun L, Chen ZJ (2013) Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341(6152):1390–1394
Kato K, Ishii R, Goto E, Ishitani R, Tokunaga F, Nureki O (2013) Structural and functional analyses of DNA-sensing and immune activation by human cGAS. PLoS ONE 8(10):e76983
Chen Q, Sun L, Chen ZJ (2016) Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat Immunol 17(10):1142
Ma Z, Ni G, Damania B (2018) Innate sensing of DNA virus genomes. Annu Rev Virol 5:341–362
Ishikawa H, Barber GN (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455(7213):674–678
Zhong B, Yang Y, Li S, Wang Y-Y, Li Y, Diao F, Lei C, He X, Zhang L, Tien P (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29(4):538–550
Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC (2008) MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol 28(16):5014–5026
Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci 106(21):8653–8658
He L, Xiao X, Yang X, Zhang Z, Wu L, Liu Z (2017) STING signaling in tumorigenesis and cancer therapy: a friend or foe? Cancer Lett 402:203–212
Maelfait J, Rehwinkel J (2017) RECONsidering sensing of cyclic dinucleotides. Immunity 46(3):337–339
Danilchanka O, Mekalanos JJ (2013) Cyclic dinucleotides and the innate immune response. Cell 154(5):962–970
Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN (2013) STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell 50(1):5–15
Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478(7370):515–518
Ishikawa H, Ma Z, Barber GN (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461(7265):788
Ablasser A, Chen ZJ (2019) cGAS in action: expanding roles in immunity and inflammation. Science 363(6431):eaat8657
Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB, Gonzalez-Dosal R, Rasmussen SB, Christensen MH, Yarovinsky TO (2012) Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol 13(8):737
Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L (2013) Structure-function analysis of STING activation by c [G (2′, 5′) pA (3′, 5′) p] and targeting by antiviral DMXAA. Cell 154(4):748–762
Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ (2013) Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell 51(2):226–235
Gao D, Li T, Li X-D, Chen X, Li Q-Z, Wight-Carter M, Chen ZJ (2015) Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci 112(42):E5699–E5705
Ahn J, Ruiz P, Barber GN (2014) Intrinsic self-DNA triggers inflammatory disease dependent on STING. J Immunol 193(9):4634–4642
Gao D, Wu J, Wu Y-T, Du F, Aroh C, Yan N, Sun L, Chen ZJ (2013) Cyclic GMP–AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341(6148):903–906
Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V (2014) Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505(7485):691
Lio C-WJ, McDonald B, Takahashi M, Dhanwani R, Sharma N, Huang J, Pham E, Benedict CA, Sharma S (2016) cGAS–STING signaling regulates initial innate control of cytomegalovirus infection. J Virol 90(17):7789–7797
Reinert LS, Lopušná K, Winther H, Sun C, Thomsen MK, Nandakumar R, Mogensen TH, Meyer M, Vægter C, Nyengaard JR (2016) Sensing of HSV-1 by the cGAS–STING pathway in microglia orchestrates antiviral defence in the CNS. Nat Commun 7:13348
Marriott I, Burmeister AR (2018) The Interleukin-10 family of cytokines and their role in the CNS. Front Cell Neurosci 12:458
Reinert LS, Rashidi AS, Tran DN, Katzilieris-Petras G, Hvidt AK, Gohr M, Fruhwürth S, Bodda C, Thomsen MK, Vendelbo MH (2021) Brain immune cells undergo cGAS/STING-dependent apoptosis during herpes simplex virus type 1 infection to limit type I IFN production. J Clin Investig 131(1):1–17
Hernáez B, Alonso G, Georgana I, El-Jesr M, Martín R, Shair KH, Fischer C, Sauer S, de Motes CM, Alcamí A (2020) Viral cGAMP nuclease reveals the essential role of DNA sensing in protection against acute lethal virus infection. Sci Adv 6(38):eaab4565
García-Belmonte R, Pérez-Núñez D, Pittau M, Richt JA, Revilla Y (2019) African swine fever virus Armenia/07 virulent strain controls interferon beta production through the cGAS–STING pathway. J Virol 93(12):e02298-e12218
He J, Hao R, Liu D, Liu X, Wu S, Guo S, Wang Y, Tien P, Guo D (2016) Inhibition of hepatitis B virus replication by activation of the cGAS–STING pathway. J Gen Virol 97(12):3368–3378
Anghelina D, Lam E, Falck-Pedersen E (2016) Diminished innate antiviral response to adenovirus vectors in cGAS/STING-deficient mice minimally impacts adaptive immunity. J Virol 90(13):5915–5927
Wang Z, Chen J, Hu J, Zhang H, Xu F, He W, Wang X, Li M, Lu W, Zeng G (2019) cGAS/STING axis mediates a topoisomerase II inhibitor-induced tumor immunogenicity. J Clin Investig 129:4850–4862
Xiong M, Wang S, Wang Y-Y, Ran Y (2018) The regulation of cGAS. Virologica Sinica 33(2):117–124
Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, Stetson DB (2012) Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med 209(11):1969–1983
Briggs LJ, Johnstone RW, Elliot RM, Xiao C-Y, Dawson M, Trapani JA, Jans DA (2001) Novel properties of the protein kinase CK2-site-regulated nuclear-localization sequence of the interferon-induced nuclear factor IFI 16. Biochem J 353(1):69–77
Veeranki S, Choubey D (2012) Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization. Mol Immunol 49(4):567–571
Dell’Oste V, Gatti D, Giorgio AG, Gariglio M, Landolfo S, De Andrea M (2015) The interferon-inducible DNA-sensor protein IFI16: a key player in the antiviral response. New Microbiol 38(1):5
Jakobsen MR, Paludan SR (2014) IFI16: at the interphase between innate DNA sensing and genome regulation. Cytokine Growth Factor Rev 25(6):649–655
Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW (2012) Structures of the HIN domain: DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36(4):561–571
Liu Z, Zheng X, Wang Y, Song H (2014) Bacterial expression of the HINab domain of IFI16: Purification, characterization of DNA binding activity, and co-crystallization with viral dsDNA. Protein Expr Purif 102:13–19
Horan KA, Hansen K, Jakobsen MR, Holm CK, Søby S, Unterholzner L, Thompson M, West JA, Iversen MB, Rasmussen SB (2013) Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol 190(5):2311–2319
Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, Jin T, Laustsen A, Hansen K, Østergaard L, Fitzgerald KA (2013) IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci 110(48):E4571–E4580
Berg RK, Rahbek SH, Kofod-Olsen E, Holm CK, Melchjorsen J, Jensen DG, Hansen AL, Jørgensen LB, Ostergaard L, Tolstrup M (2014) T cells detect intracellular DNA but fail to induce type I IFN responses: implications for restriction of HIV replication. PLoS ONE 9(1):e84513
Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC (2014) IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343(6169):428–432
Johnson KE, Chikoti L, Chandran B (2013) Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol 87(9):5005–5018
Sohn J, Morrone S, Wang T, Hooy R (2015) The cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Biophys J 108(2):40a
Ansari MA, Dutta S, Veettil MV, Dutta D, Iqbal J, Kumar B, Roy A, Chikoti L, Singh VV, Chandran B (2015) Herpesvirus genome recognition induced acetylation of nuclear IFI16 is essential for its cytoplasmic translocation, inflammasome and IFN-β responses. PLoS Pathog 11(7):e1005019
Dutta D, Dutta S, Veettil MV, Roy A, Ansari MA, Iqbal J, Chikoti L, Kumar B, Johnson KE, Chandran B (2015) BRCA1 regulates IFI16 mediated nuclear innate sensing of herpes viral DNA and subsequent induction of the innate inflammasome and interferon-β responses. PLoS Pathog 11(6):e1005030
Kobiyama K, Takeshita F, Jounai N, Sakaue-Sawano A, Miyawaki A, Ishii KJ, Kawai T, Sasaki S, Hirano H, Ishii N (2010) Extrachromosomal histone H2B mediates innate antiviral immune responses induced by intracellular double-stranded DNA. J Virol 84(2):822–832
Iqbal J, Ansari MA, Kumar B, Dutta D, Roy A, Chikoti L, Pisano G, Dutta S, Vahedi S, Veettil MV (2016) Histone H2B-IFI16 recognition of nuclear herpesviral genome induces cytoplasmic interferon-β responses. PLoS Pathog 12(10):e1005967
Li Y-G, Siripanyaphinyo U, Tumkosit U, Noranate N, Atchareeya A, Pan Y, Kameoka M, Kurosu T, Ikuta K, Takeda N (2012) Poly (I: C), an agonist of toll-like receptor-3, inhibits replication of the Chikungunya virus in BEAS-2B cells. Virol J 9(1):114
Jensen S, Thomsen AR (2012) Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 86(6):2900–2910
Zevini A, Olagnier D, Hiscott J (2017) Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol 38(3):194–205
Loo Y-M, Gale M Jr (2011) Immune signaling by RIG-I-like receptors. Immunity 34(5):680–692
Hou F, Sun L, Zheng H, Skaug B, Jiang Q-X, Chen ZJ (2011) MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146(3):448–461
Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, e Sousa CR (2006) RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314(5801):997–1001
Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, Yoneyama M, Horiuchi M, Ogura K, Fujita T (2009) Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem 284(26):17465–17474
Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, de la Cruz MJ, Borkowski B, Zhang X, Chen ZJ (2015) Correction: structural basis for the prion-like MAVS filaments in antiviral innate immunity. Elife 4:e07546
Reikine S, Nguyen JB, Modis Y (2014) Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front Immunol 5:342
Choi MK, Wang Z, Ban T, Yanai H, Lu Y, Koshiba R, Nakaima Y, Hangai S, Savitsky D, Nakasato M (2009) A selective contribution of the RIG-I-like receptor pathway to type I interferon responses activated by cytosolic DNA. Proc Natl Acad Sci 106(42):17870–17875
Zeng W, Chen ZJ (2008) MITAgating viral infection. Immunity 29(4):513–515
Ding Q, Cao X, Lu J, Huang B, Liu Y-J, Kato N, Shu H-B, Zhong J (2013) Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol 59(1):52–58
Pattabhi S, Wilkins CR, Dong R, Knoll ML, Posakony J, Kaiser S, Mire CE, Wang ML, Ireton RC, Geisbert TW (2016) Targeting innate immunity for antiviral therapy through small molecule agonists of the RLR pathway. J Virol 90(5):2372–2387
Sun B, Sundström KB, Chew JJ, Bist P, Gan ES, Tan HC, Goh KC, Chawla T, Tang CK, Ooi EE (2017) Dengue virus activates cGAS through the release of mitochondrial DNA. Sci Rep 7(1):3594
Franz KM, Neidermyer WJ, Tan Y-J, Whelan SP, Kagan JC (2018) STING-dependent translation inhibition restricts RNA virus replication. Proc Natl Acad Sci 115(9):E2058–E2067
Nitta S, Sakamoto N, Nakagawa M, Kakinuma S, Mishima K, Kusano-Kitazume A, Kiyohashi K, Murakawa M, Nishimura-Sakurai Y, Azuma S (2013) Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 57(1):46–58
Yu C-Y, Chang T-H, Liang J-J, Chiang R-L, Lee Y-L, Liao C-L, Lin Y-L (2012) Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog 8(6):e1002780
Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V (2012) DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 8(10):e1002934
Parker MT, Gopinath S, Perez CE, Linehan MM, Crawford J, Iwasaki A, Lindenbach BD (2018) Innate Immune priming by cGAS as a preparatory countermeasure against RNA virus infection. bioRxiv:434027
Su C, Zheng C (2017) Herpes simplex virus 1 abrogates the cGAS/STING-mediated cytosolic DNA-sensingpathway via its virion host shutoff protein, UL41. J Virol 91(6):e02414–02416
Biolatti M, Dell'Oste V, Pautasso S, Gugliesi F, von Einem J, Krapp C, Jakobsen MR, Borgogna C, Gariglio M, De Andrea M (2018) Human cytomegalovirus tegument protein pp65 (pUL83) dampens type I interferon productionby inactivating the DNA sensor cGAS without affecting STING. J Virol 92(6):e01774–01717
Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, Fredericks AC, Tripathi S, Zhu T, Pintado-Silva J (2017) Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNAsensing during infection. Nature Microbiol 2(5):17037
Ghosh A, Shao L, Sampath P, Zhao B, Patel NV, Zhu J, Behl B, Parise RA, Beumer JH, O’Sullivan RJ (2019) Oligoadenylate-Synthetase-Family Protein OASL Inhibits Activity of the DNA Sensor cGAS during DNA VirusInfection to Limit Interferon Production. Immunity 50(1):51–63
Kalamvoki M, Roizman B (2014) HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. In: Proceedings of the National Academy of Sciences 111 (5):E611-E617
Liu Y, Li J, Chen J, Li Y, Wang W, Du X, Song W, Zhang W, Lin L, Yuan Z (2015) Hepatitis B viruspolymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J Virol 89(4):2287–2300
Stack J, Haga IR, Schröder M, Bartlett NW, Maloney G, Reading PC, Fitzgerald KA, Smith GL, Bowie AG (2005) Vaccinia virus protein A46R targets multiple Toll-like–interleukin-1 receptor adaptors and contributes tovirulence. J Experiment Med 201(6):1007–1018
Zhang G, Chan B, Samarina N, Abere B, Weidner-Glunde M, Buch A, Pich A, Brinkmann MM, Schulz TF (2016) Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. In: Proceedings of the National Academy of Sciences 113 (8):E1034-E1043
Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B (2015) Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. In: Proceedings of the National Academy of Sciences 112 (31):E4306-E4315
Acknowledgements
We acknowledge the support of Ramalingaswami Fellowship Grant (BT/RLF/Re-entry/09/2015), Government of India, Ministry of Science and Technology, Department of Biotechnology (DBT) and Early Career Research Award Grant (File No. ECR/2018/002114) from the Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India, to author Jawed Iqbal. We also acknowledge the support of a non-NET fellowship from the Union Grant Commission (UGC) to Saleem Anwar and Khursheed Ul Islam, and a Senior Research Fellowship from ICMR, Government of India, to Md Iqbal Azmi.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Additional information
Handling Editor: Carolina Scagnolari.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Anwar, S., Ul Islam, K., Azmi, M.I. et al. cGAS–STING-mediated sensing pathways in DNA and RNA virus infections: crosstalk with other sensing pathways. Arch Virol 166, 3255–3268 (2021). https://doi.org/10.1007/s00705-021-05211-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05211-x