Abstract
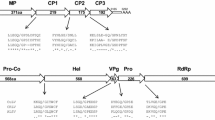
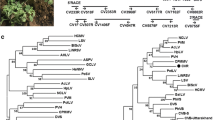
The genome of a new carlavirus isolate from asymptomatic wild Capparis spinosa L. plants in Sicily was sequenced via high-throughput sequencing (HTS) and 5’/3’ RACE experiments. The complete genomic sequence was found to be 8,280 nt in length, excluding the poly(A) tail, and contained five putative open reading frames (ORFs). Molecular characterization revealed a close relationship to caper latent virus (CapLV), with 87% and 90% nucleotide sequence identity to available partial sequences of the ORFs encoding the replicase and coat protein of that virus. According to the molecular criteria for species demarcation, which is based on the ORF-1- and ORF-5-encoded proteins, the virus characterized in this study could be considered a variant of CapLV, and we have thus designated it as CapLV-W.

Similar content being viewed by others
References
Adams MJ, Candresse T, Hammond J, Kreuze JF, Martelli GP, Namba S, Pearson MN, Ryu KH, Saldarelli P, Oshikawa N (2012) Family Betaflexiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Academic Press/Elsevier, London, pp 920–941
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Di Franco A, Gallitelli D (1987) Characterization of caper latent virus. J Phytopathol 119(2):97–105
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29(7):644–652. https://doi.org/10.1038/nbt.1883
Martin DP, Murrell B, Golden M (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. https://doi.org/10.1093/ve/vev00
Massart D, Olmos S, Jijakli H, Candresse T (2014) Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res 188:90–96
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739.
Tiberini A, Tomassoli L (2007) Caper latent virus (CapLV) partial nucleotide sequence analysis within Carlavirus group. J Plant Pathol 89(3):S63
Tiberini A, Mascia T, Tomassoli L (2011) First nucleotide sequence of a Carlavirus virus genus infecting caper. Phytopathol Mediterr 50:482–488
Tomassoli L, Tiberini A (2006) Molecular detection of caper viruses. J Plant Pathol 88(supplement 3):60
Wang R, Dong J, Wang Z, Zhou T, Li Y, Ding W (2018) Complete nucleotide sequence of a new carlavirus in chrysanthemums in China. Adv Virol 163:1973–1976
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants or animals
This study did not include experiments with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Sead Sabanadzovic.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tiberini, A., Fontana, I., Mercati, F. et al. Complete genome sequence of a new isolate of caper latent virus in caper. Arch Virol 166, 2619–2621 (2021). https://doi.org/10.1007/s00705-021-05146-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05146-3