Abstract
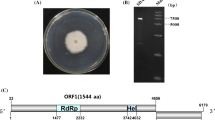
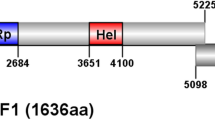
A novel mycovirus belonging to the proposed family “Fusariviridae” was discovered in Alternaria solani by sequencing a cDNA corresponding to double-stranded RNA extracted from this phytopathogenic fungus. The virus was tentatively named “Alternaria solani fusarivirus 1” (AsFV1). AsFV1 has a single-stranded positive-sense (+ssRNA) genome of 6845 nucleotides containing three open reading frames (ORFs) and a poly(A) tail. The largest ORF, ORF1, encodes a large polypeptide of 1,556 amino acids (aa) with conserved RNA-dependent RNA polymerase and helicase domains. The ORF2 and ORF3 have overlapping regions, encoding a putative protein of 522 amino acids (aa) and a putative protein of 105 amino acids (aa), respectively, both of unknown function. A multiple sequence alignment and phylogenetic analysis revealed that AsFV1 could be a new member of the “Fusariviridae”. This is the first report of the full-length nucleotide sequence of a fusarivirus that infects Alternaria solani.


Similar content being viewed by others
References
Ghabrial SA, Suzuki N (2009) Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384
Lin Y, Zhang H, Zhao C, Liu S, Guo L (2015) The complete genome sequence of a novel mycovirus from Alternaria longipes strain HN28. Arch Virol 160:577–580
Okada R, Ichinose S, Takeshita K, Urayama SI, Fukuhara T, Komatsu K et al (2018) Molecular characterization of a novel mycovirus in Alternaria alternata manifesting two-sided effects: down-regulation of host growth and up-regulation of host plant pathogenicity. Virology 519:23–32
da Silva Xavier A, de Barros APO, Godinho MT, Zerbini FM, de Oliveira Souza F, Bruckner FP, Alfenas-Zerbini P (2018) A novel mycovirus associated to Alternaria alternata comprises a distinct lineage in Partitiviridae. Virus Res 244:21–26
Hu Z, Guo J, Da Gao B, Zhong J (2020) A novel mycovirus isolated from the plant-pathogenic fungus Alternaria dianthicola. Arch Virol 165:2105–2109
Komatsu K, Katayama Y, Omatsu T, Mizutani T, Fukuhara T, Kodama M et al (2016) Genome sequence of a novel victorivirus identified in the phytopathogenic fungus Alternaria arborescens. Arch Virol 161:1701–1704
Li H, Bian R, Liu Q, Yang L, Pang T, Salaipeth L et al (2019) Identification of a novel hypovirulence-inducing hypovirus from Alternaria alternata. Front Microbiol 10:1076
Zhong J, Shang HH, Zhu CX, Zhu JZ, Zhu HJ, Hu Y, Da Gao B (2016) Characterization of a novel single-stranded RNA virus, closely related to fusariviruses, infecting the plant pathogenic fungus Alternaria brassicicola. Virus Res 217:1–7
Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479:356–368
Pearson MN, Beever RE, Boine B, Arthur K (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10:115–128
Nuss DL (1992) Biological control of chestnut blight: an example of virus-mediated attenuation of fungal pathogenesis. Microbiol Rev 56:561–576
Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G et al (2010) A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci USA 107:8387–8392
Yang D, Wu M, Zhang J, Chen W, Li G, Yang L (2018) Sclerotinia minor endornavirus 1, a novel pathogenicity debilitation-associated mycovirus with a wide spectrum of horizontal transmissibility. Viruses 10:589
Chu YM, Jeon JJ, Yea SJ, Kim YH, Yun SH, Lee YW, Kim KH (2002) Double-stranded RNA mycovirus from Fusarium graminearum. Appl Environ Microbiol 68:2529–2534
Zhang R, Liu S, Chiba S, Kondo H, Kanematsu S, Suzuki N (2014) A novel single-stranded RNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix, with similarity to hypo-like viruses. Front Microbiol 5:360
Liu R, Cheng J, Fu Y, Jiang D, Xie J (2015) Molecular characterization of a novel positive-sense, single-stranded RNA mycovirus infecting the plant pathogenic fungus Sclerotinia sclerotiorum. Viruses 7:2470–2484
Morris TJ, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Yang M, Zhai L, Xiao F, Guo Y, Fu M, Hong N, Wang G (2019) Characterization of a novel victorivirus isolated from the phytopathogenic fungus Botryosphaeria dothidea. Arch Virol 164:1609–1617
Thompson JD, Gibson TJ, Plewniak F (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Nicholas KB (1997) GeneDoc: analysis and visualization of genetic variation. Embnew News 4:14
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Acknowledgements
This study was supported by the Scientific Research Fund of the Hunan Provincial Education Department (19K040).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflicts of interest.
Ethical approval
This study did not include experiments with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Robert H.A. Coutts.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gong, W., Liu, H., Zhu, X. et al. Molecular characterization of a novel fusarivirus infecting the plant-pathogenic fungus Alternaria solani. Arch Virol 166, 2063–2067 (2021). https://doi.org/10.1007/s00705-021-05105-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05105-y