Abstract
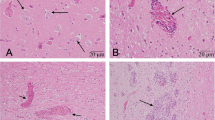
Porcine teschovirus (PTV) is a causative agent of reproductive disorders, encephalomyelitis, respiratory diseases, and diarrhea in swine, with a worldwide distribution. In this work, we identified PTV-associated nonsuppurative encephalitis as a potential cause of posterior paralysis in neonatal pigs in northeast China. Using indirect immunofluorescence assay, western blot, electron microscopy, and genome sequencing, we identified a neurotropic PTV strain, named CHN-NP1-2016, in the supernatants of pooled cerebrum and cerebellum samples from an affected piglet. Nucleotide sequence alignment revealed that the whole genome of CHN-NP1-2016 shared the highest sequence similarity (86.76% identity) with PTV 1 strain Talfan. A combination of phylogenetic and genetic divergence analysis was applied based on the deduced amino acid sequence of the P1 gene with a cutoff value of the genetic distance (0.102 ± 0.008) for defining PTV genotypes, and this showed that CHN-NP1-2016 is a variant of genotype 1. In total, 16 unique mutations and five mutant clusters were detected in the capsid proteins VP1 and VP2 of CHN-NP1-2016 when compared to other PTV1 isolates. Importantly, we detected three mutant clusters located in the exposed surface loops of the capsid protein, potentially indicating significant differences in major neutralization epitopes. Moreover, a potential recombination event in the P1 region of PTV CHN-NP1-2016 was detected. These findings provide valuable insights into the role of recombination in the evolution of teschoviruses. To our knowledge, this is the first case report of PTV-1-associated encephalitis in northeast China. Future investigations will narrow on the serology and pathogenicity of this novel isolate.








Similar content being viewed by others
References
Zell R, Delwart E, Gorbalenya AE, Hovi T, King AMQ, Knowles NJ, Lindberg AM, Pallansch MA, Palmenberg AC, Reuter G, Simmonds P, Skern T, Stanway G, Yamashita T, Ictv Report C (2017) ICTV virus taxonomy profile: picornaviridae. J Gener Virol 98:2421–2422
Zell R, Dauber M, Krumbholz A, Henke A, Birch-Hirschfeld E, Stelzner A, Prager D, Wurm R (2001) Porcine teschoviruses comprise at least eleven distinct serotypes: molecular and evolutionary aspects. J Virol 75:1620–1631
Cano-Gómez C, Fernández-Pinero J, García-Casado MA, Zell R, Jiménez-Clavero MA (2017) Characterization of PTV-12, a newly described porcine teschovirus serotype: in vivo infection and cross-protection studies. J Gener Virol 98:1636–1645
Boros Á, Nemes C, Pankovics P, Kapusinszky B, Delwart E, Reuter G (2012) Porcine teschovirus in wild boars in Hungary. Adv Virol 157:1573–1578
Yang T, Lu Y, Zhang L, Li X, Chang Y (2020) Novel species of Teschovirus B comprises at least three distinct evolutionary genotypes. Transbound Emerg Dis 67:1015–1018
Genus: Teschovirus (2019) https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/picornavirales/w/picornaviridae/704/genus-teschovirus
Yang T, Yu X, Luo B, Yan M, Li R, Qu T, Ren X (2018) Epidemiology and molecular characterization of Porcine teschovirus in Hunan, China. Transbound Emerg Dis 65:480–490
Oba M, Naoi Y, Ito M, Masuda T, Katayama Y, Sakaguchi S, Omatsu T, Furuya T, Yamasato H, Sunaga F, Makino S, Mizutani T, Nagai M (2018) Metagenomic identification and sequence analysis of a Teschovirus A-related virus in porcine feces in Japan, 2014–2016. Infect Genet Evol 66:210–216
Yang T, Lu Y, Zhang L, Li X (2020) Identification of novel genotypes belonging to the species Teschovirus A from indigenous pigs in western Jiangxi, China. Arch Virol 165:993–1001
Knowles NJ (2019) Picornaviruses. Dis Swine 2019:668–673
Deng MY, Millien M, Jacques-Simon R, Flanagan JK, Bracht AJ, Carrillo C, Barrette RW, Fabian A, Mohamed F, Moran K, Rowland J, Swenson SL, Jenkins-Moore M, Koster L, Thomsen BV, Mayr G, Pyburn D, Morales P, Shaw J, Burrage T, White W, McIntosh MT, Metwally S (2012) Diagnosis of Porcine teschovirus encephalomyelitis in the Republic of Haiti. J Vet Diagn Invest 24:671–678
Salles MW, Scholes SF, Dauber M, Strebelow G, Wojnarowicz C, Hassard L, Acton AC, Bollinger TK (2011) Porcine teschovirus polioencephalomyelitis in western Canada. J Vet Diagn Invest 23:367–373
Lin W, Cui S, Zell R (2012) Phylogeny and evolution of porcine teschovirus 8 isolated from pigs in China with reproductive failure. Arch Virol 157:1387–1391
Zhang CF, Cui SJ, Hu S, Zhang Z, Guo Q, Zell R (2010) Isolation and characterization of the first Chinese strain of porcine Teschovirus-8. J Virol Methods 167:208–213
La Rosa G, Muscillo M, Di Grazia A, Fontana S, Iaconelli M, Tollis M (2006) Validation of rt-PCR assays for molecular characterization of porcine teschoviruses and enteroviruses. J Vet Med B Infect Dis Vet Public Health 53:257–265
Cano-Gómez C, Palero F, Buitrago MD, García-Casado MA, Fernández-Pinero J, Fernández-Pacheco P, Agüero M, Gómez-Tejedor C, Jiménez-Clavero M (2011) Analyzing the genetic diversity of teschoviruses in Spanish pig populations using complete VP1 sequences. Infect Genet Evol 11:2144–2150
Sun H, Gao H, Chen M, Lan D, Hua X, Wang C, Yuan C, Yang Z, Cui L (2015) New serotypes of porcine teschovirus identified in Shanghai, China. Arch Virol 160:831–835
Yang T, Li R, Yao Q, Zhou X, Liao H, Ge M, Yu X (2018) Prevalence of Porcine teschovirus genotypes in Hunan, China: identification of novel viral species and genotypes. J Gener Virol 99:1261–1267
Wang B, Tian ZJ, Gong DQ, Li DY, Wang Y, Chen JZ, An TQ, Peng JM, Tong GZ (2010) Isolation of serotype 2 porcine teschovirus in China: evidence of natural recombination. Vet Microbiol 146:138–143
Zhang MJ, Liu DJ, Liu XL, Ge XY, Jongkaewwattana A, He QG, Luo R (2019) Genomic characterization and pathogenicity of porcine deltacoronavirus strain CHN-HG-2017 from China. Adv Virol 164:413–425
Reed LJ, Muench H (1938) A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497
Schubert M, Lindgreen S, Orlando L (2016) AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes 9:88
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1:18
Li H, Ruan J, Durbin R (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18:1851–1858
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174-181
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003
Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160
Kaku Y, Murakami Y, Sarai A, Wang Y, Ohashi S, Sakamoto K (2007) Antigenic properties of porcine teschovirus 1 (PTV-1) Talfan strain and molecular strategy for serotyping of PTVs. Adv Virol 152:929–940
Mateu MG (1995) Antibody recognition of picornaviruses and escape from neutralization: a structural view. Virus Res 38:1–24
Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG (2012) Crystal structure of human enterovirus 71. Sci (N Y) 336:1274
Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG et al (1985) Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145–153
Foo DG, Alonso S, Phoon MC, Ramachandran NP, Chow VT, Poh CL (2007) Identification of neutralizing linear epitopes from the VP1 capsid protein of Enterovirus 71 using synthetic peptides. Virus Res 125:61–68
Simmonds P (2006) Recombination and selection in the evolution of picornaviruses and other Mammalian positive-stranded RNA viruses. J Virol 80:11124–11140
Saeng-Chuto K, Stott CJ, Wegner M, Kaewprommal P, Piriyapongsa J, Nilubol D (2018) The full-length genome characterization, genetic diversity and evolutionary analyses of Senecavirus A isolated in Thailand in 2016. Infect Genet Evol 64:32–45
Mandary MB, Poh CL (2018) Changes in the EV-A71 genome through recombination and spontaneous mutations: impact on virulence. Viruses 10(6):320
Oberste MS, Maher K, Pallansch MA (2004) Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J Virol 78:855–867
Ferretti L, Di Nardo A, Singer B, Lasecka-Dykes L, Logan G, Wright CF, Pérez-Martín E, King DP, Tuthill TJ, Ribeca P (2018) Within-host recombination in the foot-and-mouth disease virus genome. Viruses 10(5):221
Feng L, Shi HY, Liu SW, Wu BP, Chen JF, Sun DB, Tong YE, Fu MS, Wang YF, Tong GZ (2007) Isolation and molecular characterization of a porcine teschovirus 1 isolate from China. Acta Virol 51:7–11
Acknowledgements
This work was supported by China Agriculture Research System (No. CARS-35).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Additional information
Handling Editor: Ana Cristina Bratanich.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
705_2021_4994_MOESM7_ESM.docx
Table S3 Estimates of evolutionary divergence over sequence pairs within and between PTV genotypes, including VP1 sequences of CHN-NP1-2016 and 148 PTV reference strains (mean ± SE) (DOCX 26 KB)
705_2021_4994_MOESM8_ESM.docx
Table S4 Estimates of evolutionary divergence over sequence pairs within and between PTV genotypes, including VP2 sequences of CHN-NP1-2016 and 133 PTV reference strains (mean ± SE) (DOCX 26 KB)
Rights and permissions
About this article
Cite this article
Ma, H., Zhang, M., Wu, M. et al. Isolation and genetic characteristics of a neurotropic teschovirus variant belonging to genotype 1 in northeast China. Arch Virol 166, 1355–1370 (2021). https://doi.org/10.1007/s00705-021-04994-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-04994-3