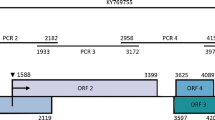
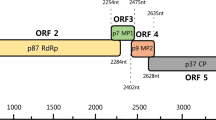
Abstract
Ornithogalum thyrsoides, a widely cultivated bulbous ornamental plant endemic to South Africa, has significant commercial value as a pot plant and for the production of cut flowers. However, infection by viruses threatens the success of commercial cultivation, as symptoms negatively affect the appearance of the plant and flowers. To date, four Ornithogalum-infecting viruses have been reported. Complete genome sequence data are available for three of these viruses, but the genome of the potyvirus ornithogalum virus 3 (OV3) has not been fully sequenced. In this study, the complete sequence of OV3 was determined by high-throughput sequencing (HTS) and validated by Sanger sequencing. Based on recognition of protease cleavage patterns and multiple sequence alignments with closely related viruses, the polyprotein of OV3 was predicted to be proteolytically cleaved to produce 10 mature peptides containing domains conserved in members of the genus Potyvirus. Phylogenetic analysis and species demarcation criteria confirm the previous classification of OV3 as a member of a separate species in this genus. This is the first report of a complete genome sequence of OV3.


Similar content being viewed by others
References
Bodkin F (1986) Encyclopaedia botanica: the essential reference guide to native and exotic plants in Australia. Angus & Robertson, Sydney
Mostert I, Visser M, Gazendam I et al (2020) Complete genome sequence of a novel polerovirus in Ornithogalum thyrsoides from South Africa. Arch Virol 165:483–486. https://doi.org/10.1007/s00705-019-04472-x
Burger JT, von Wechmar MB (1989) Purification and some properties of South African isolates of Ornithogalum mosaic virus. Phytopathology 79:385–391. https://doi.org/10.1094/Phyto-79-385
Matsumoto T, Yamamoto H, Fuji S, Inoue M (2007) Isolation and characterization of a novel potyvirus tentatively named Ornithogalum virus 2. J Gen Plant Pathol 73:222–224. https://doi.org/10.1007/s10327-007-0010-3
Matsumoto T, Yamamoto H, Fuji S, Inoue M (2008) Characterization of a novel potyvirus tentatively named Ornithogalum virus 3, successfully isolated from O. thyrsoides co-infected with two other potyviruses by single-aphid inoculation. J Gen Plant Pathol 74:76–80. https://doi.org/10.1007/s10327-007-0057-1
Adams M, Antoniw J, Beaudoin F (2005) Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol Plant Pathol 6:471–487. https://doi.org/10.1111/j.1364-3703.2005.00296.x
Chung BYW, Miller WA, Atkins JF, Firth AE (2008) An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci USA 105:5897–5902. https://doi.org/10.1073/pnas.0800468105
Shiboleth YM, Haronsky E, Leibman D et al (2007) The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J Virol 81:13135–13148. https://doi.org/10.1128/JVI.01031-07
Revers F, García JA (2015) Chapter three—molecular biology of potyviruses. In: Maramorosch K, Mettenleiter TC (eds) Advances in virus research. Academic Press, pp 101–199
Atreya CD, Raccah B, Pirone TP (1990) A point mutation in the coat protein abolishes aphid transmissibility of a potyvirus. Virology 178:161–165. https://doi.org/10.1016/0042-6822(90)90389-9
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. MolBiolEvol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Adams MJ, Antoniw JF, Fauquet CM (2005) Molecular criteria for genus and species discrimination within the family Potyviridae. Arch Virol 150:459–479. https://doi.org/10.1007/s00705-004-0440-6
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Rachelle Bester, Ilani Mostert, Dirk Aldrich, and Marike Visser. The first draft of the manuscript was written by Ilani Mostert, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare they have no conflict of interest.
Ethical approval
This article does not contain any studies involving human participants or animals.
Additional information
Handling Editor: Ioannis E. Tzanetakis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mostert, I., Bester, R., Aldrich, D. et al. Complete genome sequence of ornithogalum virus 3. Arch Virol 166, 1213–1216 (2021). https://doi.org/10.1007/s00705-021-04965-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-04965-8