Abstract
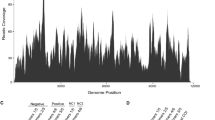
The leaf beetle Aulacophora lewisii (family Chrysomelidae, order Coleoptera) is a common insect pest of cucurbitaceous vegetables. In this study, the complete genome sequence of a novel virus from a single leaf beetle was determined using metagenomic sequencing and rapid amplification of cDNA ends. A homology search and phylogenetic analysis suggested that the new virus belongs to the genus Iflavirus, family Iflaviridae, and it was tentatively named “Aulacophora lewisii iflavirus 1” (ALIV1). ALIV1 has a single positive-stranded RNA genome of 9655 nucleotides in length (excluding the polyA tail) that is predicted to encode typical conserved domains of iflaviruses, including two picornavirus-like capsid protein domains, a helicase domain, and an RNA-dependent RNA polymerase (RdRp) domain. Sequence comparisons showed that the full genome sequence of ALIV1 is most similar to that of Brevicoryne brassicae picorna-like virus, with 42.4% sequence identity, and it shares 60% sequence identity in the coat protein region with its closest homolog, Watson virus. The average coverage of the ALIV1 sequence was approximately 5000X, suggesting that it might actively replicate in the host. Phylogenetic analysis based on deduced amino acid sequences suggested that ALIV1 is closely related to Dinocampus coccinellae paralysis virus. To the best of our knowledge, ALIV1 is the first virus discovered in A. lewisii and is also the first iflavirus identified in a member of the genus Aulacophora.


Similar content being viewed by others
References
Valles SM, Chen Y, Firth AE, Guerin DMA, Hashimoto Y, Herrero S, de Miranda JR, Ryabov E, Ictv Report C (2017) ICTV virus taxonomy profile: Iflaviridae. J Gen Virol 98:527–528
Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, Mead A, Burroughs N, Evans DJ (2014) A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog 10:e1004230
Bailey L, Gibbs AJ, Woods RD (1964) Sacbrood virus of the larval honey bee (Apis Mellifera Linnaeus). Virology 23:425–429
Carrillo-Tripp J, Krueger EN, Harrison RL, Toth AL, Miller WA, Bonning BC (2014) Lymantria dispar iflavirus 1 (LdIV1), a new model to study iflaviral persistence in lepidopterans. J Gen Virol 95:2285–2296
Murakami R, Suetsugu Y, Kobayashi T, Nakashima N (2013) The genome sequence and transmission of an iflavirus from the brown planthopper, Nilaparvata lugens. Virus Res 176:179–187
Roundy CM, Azar SR, Rossi SL, Weaver SC, Vasilakis N (2017) Insect-specific viruses: a historical overview and recent developments. Adv Virus Res 98:119–146
Abe M, Matsuda K (2005) Chemical factors influencing the feeding preference of three Aulacophora leaf beetle species (Coleoptera: Chrysomelidae). Appl Entomol Zool 40:161–168
Liu S, Chen Y, Sappington TW, Bonning BC (2017) Genome sequence of the first coleopteran iflavirus isolated from western corn rootworm, Diabrotica virgifera virgifera LeConte. Genome Announc 2017:5
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T (2020) ModelTest-NG: a new and scalable tool for the selection of dna and protein evolutionary models. Mol Biol Evol 37:291–294
Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Funding
This research was supported by the China National Novel Transgenic Organisms Breeding Project (2019ZX08004-004) and the Ningbo Science and Technology Innovation 2025 Major Project (2019B10004). This work was sponsored by the K.C. Wong Magna Fund at Ningbo University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical statement
No experimental work with humans was done in this study.
Additional information
Handling Editor: Jesús Navas-Castillo.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2020_4859_MOESM2_ESM.tif
Fig. S1 Maximum-likelihood phylogenetic tree base on the full genome sequences of Aulacophora lewisii iflavirus 1 (ALIV1) and the other iflaviruses. Bootstrap values are shown at each node of the tree (when > 50). Scale bars represent percent divergence. The ALIV1 isolate identified in this study is indicated by a red triangle (TIF 1554 kb)
Rights and permissions
About this article
Cite this article
Ye, ZX., Li, YH., Chen, JP. et al. Complete genome analysis of a novel iflavirus from a leaf beetle, Aulacophora lewisii. Arch Virol 166, 309–312 (2021). https://doi.org/10.1007/s00705-020-04859-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04859-1