Abstract
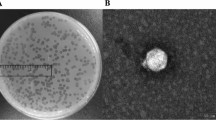
A virulent phage, named vB_EcoS_XY3, was isolated from hospital wastewater in Xiangyang, China. Its morphological characteristics, growth parameters, adsorption rate, and pH and temperature stability were determined. Phage vB_EcoS_XY3 was found to be able to infect Escherichia coli laboratory strains and also some multidrug-resistant E. coli strains. Its complete genome consists of 51,345 base pairs of double-stranded DNA with an average GC content of 55.24% and 85 putative protein-coding genes. Forty-four genes were annotated with known functions. These results will not only provide further insights into E. coli phages but also have implications for the development of potential biocontrol agents.



Similar content being viewed by others
References
Jang J, Hur HG, Sadowsky MJ, Byappanahalli MN, Yan T, Ishii S (2017) Environmental Escherichia coli: ecology and public health implications—a review. J Appl Microbiol 123(3):570–581. https://doi.org/10.1111/jam.13468
Ron EZ (2010) Distribution and evolution of virulence factors in septicemic Escherichia coli. Int J Med Microbiol 300(6):367–370. https://doi.org/10.1016/j.ijmm.2010.04.009
Wiles TJ, Kulesus RR, Mulvey MA (2008) Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol 85(1):11–19. https://doi.org/10.1016/j.yexmp.2008.03.007
Russo TA, Johnson JR (2003) Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 5(5):449–456. https://doi.org/10.1016/S1286-4579(03)00049-2
van Hout D, Verschuuren TD, Bruijning-Verhagen PCJ, Bosch T, Schürch AC, Willems RJL, Bonten MJM, Kluytmans JAJW (2020) Extended-spectrum beta-lactamase (ESBL)-producing and non-ESBL-producing Escherichia coli isolates causing bacteremia in the Netherlands (2014–2016) differ in clonal distribution, antimicrobial resistance gene and virulence gene content. PLoS One 15(1):e0227604–e0227604. https://doi.org/10.1371/journal.pone.0227604
Bloom DE, Black S, Salisbury D, Rappuoli R (2018) Antimicrobial resistance and the role of vaccines. Proc Natl Acad Sci USA 115(51):12868–12871. https://doi.org/10.1073/pnas.1717157115
Moelling K, Broecker F, Willy C (2018) A wake-up call: we need phage therapy now. Viruses. https://doi.org/10.3390/v10120688
Gu J, Liu X, Li Y, Han W, Lei L, Yang Y, Zhao H, Gao Y, Song J, Lu R, Sun C, Feng X (2012) A method for generation phage cocktail with great therapeutic potential. PLoS One 7(3):e31698. https://doi.org/10.1371/journal.pone.0031698
Govind R, Fralick JA, Rolfe RD (2006) Genomic organization and molecular characterization of Clostridium difficile bacteriophage PhiCD119. J Bacteriol 188(7):2568–2577. https://doi.org/10.1128/JB.188.7.2568-2577.2006
Kropinski AM (2018) Practical advice on the one-step growth curve. Methods Mol Biol 1681:41–47. https://doi.org/10.1007/978-1-4939-7343-9_3
Yuan Y, Gao M, Wu D, Liu P, Wu Y (2012) Genome characteristics of a novel phage from Bacillus thuringiensis showing high similarity with phage from Bacillus cereus. PLoS One 7(5):e37557. https://doi.org/10.1371/journal.pone.0037557
Patel RK, Jain M (2012) NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7(2):e30619. https://doi.org/10.1371/journal.pone.0030619
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Lowe TM, Chan PP (2016) tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44(W1):W54–W57. https://doi.org/10.1093/nar/gkw413
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35(9):3100–3108. https://doi.org/10.1093/nar/gkm160
Lee I, Ouk Kim Y, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66(2):1100–1103. https://doi.org/10.1099/ijsem.0.000760
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Stothard P, Wishart DS (2005) Circular genome visualization and exploration using CGView. Bioinformatics (Oxf, Engl) 21(4):537–539. https://doi.org/10.1093/bioinformatics/bti054
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics (Oxf, Engl) 27(7):1009–1010. https://doi.org/10.1093/bioinformatics/btr039
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31(13):3497–3500. https://doi.org/10.1093/nar/gkg500
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Murphy J, Mahony J, Ainsworth S, Nauta A, van Sinderen D (2013) Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence. Appl Environ Microbiol 79(24):7547–7555. https://doi.org/10.1128/aem.02229-13
Adriaenssens E, Brister JR (2017) How to name and classify your phage: an informal guide. Viruses. https://doi.org/10.3390/v9040070
Abel Carrias TJW, Waldbieser Geoffrey C, Mead David A, Terhune Jeffery S, Liles Mark R (2011) Comparative genomic analysis of bacteriophages specific to the channel catfish pathogen Edwardsiella ictaluri. Virol J 8(1):6. https://doi.org/10.1186/1743-422X-8-6
Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM (2014) Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52(5):1501–1510. https://doi.org/10.1128/jcm.03617-13
Nilsson AS (2014) Phage therapy—constraints and possibilities. Upsala J Med Sci 119(2):192–198. https://doi.org/10.3109/03009734.2014.902878
Acknowledgements
This investigation was supported by the Natural Science Foundation of Hubei Province of China (2020DFE025, 2019AHB068, 2018CFB701), the Scientific Research Project of Hubei Province Health Committee (ZY2019F028), the Innovative Team Project (2017YHKT02) from the Institute of Medicine and Nursing at Hubei University of Medicine and the Science and Technology Development Project of Xiangyang.
Author information
Authors and Affiliations
Contributions
PF, QZ and MS conceived and designed the experiments. PF, QZ, LS, QX, HX and SC performed the experiments. PF, QZ, HX, SC and QX analyzed the data. PF wrote the paper. PF, QZ, HX, SC, QX, XS and MS revised the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Stability of phage vB_EcoS_XY3. (A) Thermal tolerance of the phage. (B) Tolerance of the phage to treatment at different pH values. Three independent experiments were performed, and the values represented by means. (EPS 4190 kb)
Fig. S2
Kinetics of vB_EcoS_XY3 adsorption onto host cells (A) and one-step growth curve of phage vB_EcoS_XY3 (B). Data are shown as the average of triplicate experiments, and error bars indicate the standard deviation (SD). (EPS 6453 kb)
Fig. S3
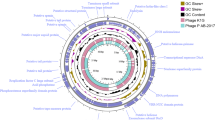
Genome structure of phage vB_EcoS_XY3. The middle ring indicates the GC content (black), and the innermost ring represents the GC skew of the vB_EcoS_XY3 genome. The predicted functions of the CDSs are indicated. (EPS 10569 kb)
Rights and permissions
About this article
Cite this article
Fu, P., Zhao, Q., Shi, L. et al. Isolation and complete genome sequencing of the virulent phage vB_EcoS_XY3 infecting multidrug-resistant Escherichia coli. Arch Virol 166, 303–307 (2021). https://doi.org/10.1007/s00705-020-04844-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04844-8