Abstract
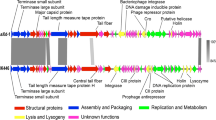
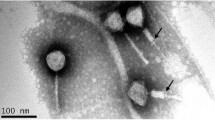
Pseudomonas phages PaGz-1 and PaZq-1, two new phages infecting Pseudomonas aeruginosa, were isolated from fresh water in Guangdong province, China. The genomes of these two phages consist of 93,975 bp and 94,315 bp and contain 175 and 172 open reading frames (ORFs), respectively. The genome sequences of PaGz-1 and PaZq-1 share 95.8% identity with a query coverage of 94%, suggesting that these two phages belong to two different species. Based on results of nucleotide sequence alignment, gene annotation, and phylogenetic analysis, we propose PaGz-1 and PaZq-1 as representative isolates of two species in the genus Pakpunavirus within the family Myoviridae.


Similar content being viewed by others
References
Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D (2018) Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health 2018:60–66
Oliveira H, Pinto G, Oliveira A, Noben JP, Hendrix H, Lavigne R, Lobocka M, Kropinski AM, Azeredo J (2017) Characterization and genomic analyses of two newly isolated Morganella phages define distant members among Tevenvirinae and Autographivirinae subfamilies. Sci Rep 7:46157
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Besemer J, Borodovsky M (2005) GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res 33((Web Server issue)):W451–W454
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics (Oxf Engl) 27(7):1009–1010
Shen M, Le S, Jin X, Li G, Tan Y, Li M, Zhao X, Shen W, Yang Y, Wang J, Zhu H, Li S, Rao X, Hu F, Lu S (2016) Characterization and comparative genomic analyses of Pseudomonas aeruginosa phage PaoP5: new members assigned to PAK_P1-like viruses. Sci Rep 6(1):34067
King AMQ, Lefkowitz EJ, Mushegian AR, Adams MJ, Dutilh BE, Gorbalenya AE, Harrach B, Harrison RL, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert ML, Rubino L, Sabanadzovic S, Sanfacon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Davison AJ (2018) Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch Virol 163:2601–2631
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Bailly-Bechet M, Vergassola M, Rocha E (2007) Causes for the intriguing presence of tRNAs in phages. Genome Res 17:1486–1495
Henry M, Bobay LM, Chevallereau A, Saussereau E, Ceyssens PJ, Debarbieux L (2015) The search for therapeutic bacteriophages uncovers one new subfamily and two new genera of Pseudomonas-infecting Myoviridae. PLoS One 10:e0117163
Acknowledgements
We are grateful to J. Feng, Institute of Microbiology of the Chinese Academy of Sciences, for providing bacterial strains. This work was supported by the National Natural Science Foundation of China (31770111, 31971350, 31800083); the Shenzhen Science Technology and Innovation Commission (JCYJ20170413153329565, JCYJ20170818160418654, JCYJ20180302145817753, JCYJ20170818155011625, KQTD2016112915000294, KQTD2015033117210153), the Engineering Laboratory for Automated Manufacturing of Therapeutic Synthetic Microbes (Shenzhen Development and Reform Commission No. [2016]1194), and the Research Instrument Development Project of the Chinese Academy of Sciences (YJKYYQ20170063).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wen, L., Chen, L., Yuan, S. et al. Complete genome analysis of PaGz-1 and PaZq-1, two novel phages belonging to the genus Pakpunavirus. Arch Virol 165, 2393–2396 (2020). https://doi.org/10.1007/s00705-020-04745-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04745-w