Abstract
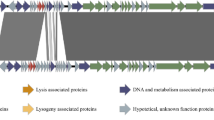
Genomic evolution among bacteriophages infecting Caulobacter crescentus is inevitable. However, the conservation of the proteins associated with intact phage particles has not been investigated. In this study, we compared the structural proteins associated with two genomically diverse but morphologically similar C. crescentus-infecting bacteriophages, phiCbK and CcrSC. We were able to detect more than 20 proteins that are part of the bacteriophage particle in both phages, and we were able to identify a small number of proteins that were found in only one of the two phage particles. All but one of the genes coding for these structural proteins were located in a region of the genome that had been designated a structural region, confirming the idea that the genes in these phage genomes are clustered according to their function. During the purification process, we also discovered that phiCbk has a replication complex that can be recovered from the cell lysate, and this complex allowed us to identify many of the phage proteins involved in phage genome replication.




Similar content being viewed by others
References
Clokie MRJ, Millard AD, Letarov AD, Heaphy S (2011) Phages in nature. Bacteriophage 1:31–45. https://doi.org/10.4161/bact.1.1.14942
Grose JH, Casjens SR (2014) Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology 468:421–443
Poindexter JS (1964) Biological properties and classification of the Caulobacter group. Bacteriol Rev 28:231–295
Agabian-Keshishian N, Shapiro L (1971) Bacterial differentiation and phage infection. Virology 44:46–53
Panis G, Lambert C, Viollier PH (2012) Complete genome sequence of Caulobacter crescentus bacteriophage φCbK. J Virol 86:10234–10235. https://doi.org/10.1128/JVI.01579-12
Leonard KR, Kleinschmidt AK, Agabian-KeshishianN SL, Maizel JV (1972) Structural studies on the capsid of Caulobacter crescentus bacteriophage PhiCbK. J Mol Biol 71:201–216. https://doi.org/10.1016/0022-2836(72)90346-4
Johnson RC, Wood NB, Ely B (1977) Isolation and characterization of bacteriophages for Caulobacter crescentus. J Gen Virol 37:323–335. https://doi.org/10.1099/0022-1317-37-2-323
Guerrero-Ferreira RC, Viollier PH, Ely B, Poindexter JS, Georgieva M, Jensen GJ, Wright ER (2011) Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc Natl Acad Sci USA 108:9963–9968. https://doi.org/10.1073/pnas.1012388108
Christen M, Beusch C, Bosch Y, Cerletti D, Flores-Tinoco C, Medico L, Tschan F, Christen B (2016) Quantitative selection analysis of bacteriophage CbK susceptibility in Caulobacter crescentus. J Mol Biol 428:419–430. https://doi.org/10.1016/j.jmb.2015.11.018
Gill JJ, Berry JD, Russell WK, Lessor L, Escobar-Garcia DA, Hernandez D et al (2012) The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genom 13:542. https://doi.org/10.1186/1471-2164-13-542
Wilson K, Ely B (2019) Analyses of four new Caulobacter Phicbkviruses indicate independent lineages. J Gen Virol 100:321–331. https://doi.org/10.1099/jgv.0.001218
Callahan CT, Wilson KM, Ely B (2015) Characterization of the proteins associated with Caulobacter crescentus bacteriophage CbK particles. Curr Microbiol 71:75–80. https://doi.org/10.1007/s00284-015-0922-7
Ash K, Drake K, Gibbs W, Ely B (2017) Genomic diversity of type B3 bacteriophages of Caulobacter crescentus. Curr Microbiol 74:779–786. https://doi.org/10.1007/s00284-017-1248-4
Ely B, Croft RH (1982) Transposon mutagenesis in Caulobacter crescentus. J Bacteriol 149:620–625
Bonilla N, Rojas M, Cruz GNF, Hung S, Rohwer F, Barr J (2016) Phage on tap-a quick and efficient protocol for the preparation of bacteriophage laboratory stocks. PeerJ. https://doi.org/10.7717/peerj.2261
Boja ES, Fales HM (2001) Overalkylation of a protein digest with iodoacetamide. Anal Chem 73(15):3576–3582. https://doi.org/10.1021/ac0103423
Nielsen ML, Vermeulen M, Bonaldi T, Cox J, Moroder L, Mann M (2008) Iodoacetamide-induced artifact mimics ubiquitination in mass spectroscopy. Nat Methods 5:459–460. https://doi.org/10.1038/nmeth0608-459
Nie L, Zhu M, Sun S, Zhai L, Wu Z, Qian L, Tan M (2016) An optimization of the LC-MS/MS workflow for deep proteome profiling on an Orbitrap Fusion. Anal Meth 8:425–434. https://doi.org/10.1039/C5AY01900A
Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75(17):4646–4658. https://doi.org/10.1021/ac0341261
Altwchul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu Y-K (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109
Chiu CS, Cook KS, Greenberg GR (1982) Characteristics of a bacteriophage T4-induced complex synthesizing deoxyribonucleotides. J Biol Chem 267:15087–15097
Funding
This work was funded in part by National Institutes of Health grant GM076277 to BE.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wilson, K., Zhu, F., Zheng, R. et al. Identification of proteins associated with two diverse Caulobacter phicbkvirus particles. Arch Virol 165, 1995–2002 (2020). https://doi.org/10.1007/s00705-020-04707-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04707-2