Abstract
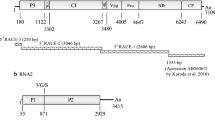
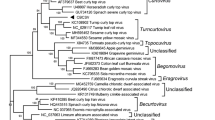
The complete genome sequence of a novel comovirus identified in Guanajuato, Mexico, in a common bean plant (Phaseolus vulgaris L.) coinfected with Phaseolus vulgaris alphaendornavirus 1 (PvEV-1) and Phaseolus vulgaris alphaendornavirus 2 (PvEV-2) is presented. According to the current ICTV taxonomic criteria, this comovirus corresponds to a new species, and the name "Phaseolus vulgaris severe mosaic virus" (PvSMV) is proposed for this virus based on the observed symptoms of “severe mosaic” syndrome caused by comoviruses in common bean. PvSMV is closely related to bean pod mosaic virus (BPMV), and its genome consists of two polyadenylated RNAs. RNA-1 (GenBank accession number MN837498) is 5969 nucleotides (nt) long and encodes a single polyprotein of 1856 amino acids (aa), with an estimated molecular weight (MW) of 210 kDa, that contains putative proteins responsible for viral replication and proteolytic processing. RNA-2 (GenBank accession number MN837499) is 3762 nt long and encodes a single polyprotein of 1024 aa, with an estimated MW of 114 kDa, that contains putative movement and coat proteins. Cleavage sites were predicted based on similarities in size and homology to aa sequences of other comoviruses available in the GenBank database. Symptoms associated with PvSMV include mosaic, local necrotic lesions, and apical necrosis. This is the first report of a comovirus infecting common bean in Mexico.


Similar content being viewed by others
References
Delgadillo-Sánchez F, Garzón-Tiznado JA, Vega-Pina A (1989) Cucurbit viruses in Mexico: a survey. Rev Mex Fitopatol 7:136–139
Rocha-Pena MA, Fulton JP (1984) Some properties of a cowpea severe mosaic virus isolate from Tabasco, Mexico. Turrialba 34(2):237–242
Wellink J, Rezelman G, Goldbach R, Beyreuther K (1986) Determination of the proteolytic processing sites in the polyprotein encoded by the bottom-competent RNA of Cowpea mosaic virus. J Virol 59:50–58
Di D, Hu CC, Ghabrial SA (1999) Complete nucleotide sequence of bean pod mottle virus RNA1: sequence comparisons and evolutionary relationships to other comoviruses. Virus Genes 18:129–137
MacFarlane SA, Shanks M, Davies JW, Zlotnick A, Lomonossoff GP (1991) Analysis of the nucleotide sequence of bean pod mottle virus middle component RNA. Virology 183:405–409
Wellink J, Van Kammen AB (1989) Cell-to-cell transport of cowpea mosaic virus requires both the 58K/48K proteins and the capsid proteins. J Gen Virol 70(9):2279–2286
Morales FJ, Castaño M (1992) Increased disease severity induced by some comoviruses in bean genotypes possessing monogenic dominant resistance to bean common mosaic potyvirus. Plant Dis 76(6):570–573
Morales FJ, Singh SP (1997) Inheritance of the mosaic and necroses reactions induced by bean severe mosaic comoviruses in Phaseolus vulgaris L. Euphytica 93(2):223–226
Chiquito-Almanza E, Acosta-Gallegos J, García-Álvarez N, Garrido-Ramírez E, Montero-Tavera V, Guevara-Olvera L et al (2017) Simultaneous detection of both RNA and DNA viruses infecting dry bean and occurrence of mixed infections by BGYMV, BCMV, and BCMNV in the Central-west region of Mexico. Viruses 9(4):63
Zheng Y, Gao S, Padmanabhan C, Li R, Galvez M, Gutierrez D et al (2017) VirusDetect: An automated pipeline for efficient virus discovery using deep sequencing of small RNAs. Virology 500:130–138
Goldbach R, Krijt J (1982) Cowpea mosaic virus-encoded protease does not recognize primary translation products of M RNAs from other comoviruses. J Virol 43(3):1151–1154
Vos P, Verver J, van Wezenbeek P, Van Kammen A, Goldbach R (1984) Study of the genetic organisation of a plant viral RNA genome by in vitro expression of a full-length DNA copy. EMBO J 3(13):3049–3053
Muhire BM, Varsani A, Martin DP (2014) SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9(9):e108277
Thompson JR, Dasgupta I, Fuchs M, Iwanami T, Karasev AV, Petrzik K et al (2017) ICTV Virus Taxonomy Profile: Secoviridae. J Gen Virol 98:529–531
Lelli D, Moreno A, Steyer A, Naglic T, Chiapponi C, Prosperi A et al (2015) Detection and characterization of a novel reassortant mammalian orthoreovirus in bats in Europe. Viruses 7(11):5844–5854
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol 7(11):5844–5854
Gu H, Clark AJ, De Sa PB, Pfeiffer TW, Tolin S, Ghabrial SA (2002) Diversity among isolates of Bean pod mottle virus. Phytopathology 92(4):446–452
Okada R, Yong CK, Valverde RA, Sabanadzovic S, Aoki N, Hotate S et al (2013) Molecular characterization of two evolutionarily distinct endornaviruses co-infecting common bean (Phaseolus vulgaris). J Gen Virol 94(1):220–229
Okada R, Alcalá-Briseño RI, Escalante C, Sabanadzovic S, Valverde RA (2018) Genomic sequence of a novel endornavirus from Phaseolus vulgaris and occurrence in mixed infections with two other endornaviruses. Virus Res 257:63–67
Khankhum S, Valverde RA (2018) Physiological traits of endornavirus-infected and endornavirus-free common bean (Phaseolus vulgaris) cv Black Turtle Soup. Arch Virol 163(4):1051–1056
Acknowledgements
This study was funded by INIFAP project no. 1551434856, and by CONACYT project no. 2146441.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Tim Skern.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chiquito-Almanza, E., Zamora-Aboytes, J.M., Medina, H.R. et al. Complete genome sequence of a novel comovirus infecting common bean. Arch Virol 165, 1505–1509 (2020). https://doi.org/10.1007/s00705-020-04610-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04610-w