Abstract
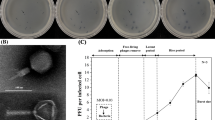
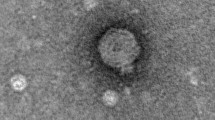
A pathogen of significance in the aquaculture sector, the Gram-negative marine bacterium Vibrio parahaemolyticus causes gastroenteritis associated with consumption of improperly prepared seafood. This bacterium can be controlled using lytic bacteriophages as an alternative to antibiotics. ϕVP-1 is a lytic phage of V. parahaemolyticus that was isolated from an aquafarm water sample with the aim of assessing its potential as a bio-control agent and determining its physicochemical properties and genomic sequence. Morphological analysis by transmission electron microscopy and phylogenetic analysis based on the large terminase subunit gene showed that this phage belongs to the family Myoviridae. It could infect multiple-drug-resistant (MDR) V. parahaemolyticus and V. alginolyticus strains of mangrove and seafood origin. With a maximum adsorption time of 30 min, ϕVP-1 has a short latent period of 10 min with burst size of 44 particles/cell. Whole-genome sequencing was done using the Illumina platform, and annotation was done using GeneMarkS and Prodigal. The 150,764bp genome with an overall G+C content of 41.84% had 203 putative protein-encoding open reading frames, one tRNA gene, and 66 predicted promoters. A number of putative DNA replication and regulation, DNA packaging and structure, and host lysis genes were identified. Comparison of the ϕVP-1 genome sequence to those of known Vibrio phages indicated little discernible DNA sequence similarity, suggesting that ϕVP-1 is a novel Vibrio phage. Sequence analysis revealed the presence of 64 potential ORFs with a T4-like genomic organization. In silico analysis suggested an obligate lytic life cycle and showed the absence of lysogeny or virulence genes. The complete sequence of ϕVP-1 was annotated and deposited in the GenBank database (accession no. MH363700). The genetic features of this novel phage suggest that it might be applicable for phage therapy against pathogenic strains of V. parahaemolyticus.







Similar content being viewed by others
References
DePaola A, Ulaszek J, Kaysner CA, Tenge BJ, Nordstrom JL, Wells J, Gendel SM (2003) Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl Environ Microbiol 69(7):3999–4005
Lee CT, Chen IT, Yang YT, Ko TP, Huang YT, Huang JY, Lightner DV (2015) The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc Natl Acad Sci 112(34):10798–10803
Golkar Z, Bagasra O, Pace DG (2014) Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. J Infect Dev Ctries 8:129–136. https://doi.org/10.3855/jidc.3573
Phumkhachorn P, Rattanachaikunsopon P (2010) Isolation and partial characterization of a bacteriophage infecting the shrimp pathogen Vibrio harveyi. Afr J Microbiol Res 4(16):1794–1800
Adams MH (1959) Bacteriophages. Wiley, New York
Maniatis T, Fritsch EF, Sambrook J (2000) Molecular cloning: a laboratory manual, vol 545. Cold Spring Harbor Laboratory, Cold Spring Harbor
Hyman P, Abedon ST (2009) Practical methods for determining phage growth paramenters. In: Clokie MRJ, Kropinski AM (eds) Bacteriophages: methods and protocols, vol 1. Isolation, characterization and interactions. Humana Press, New York, pp 175–202
Capra ML, Quiberoni ADL, Ackermann HW, Moineau S, Reinheimer JA (2006) Characterization of a new virulent phage (MLC-A) of Lactobacillus paracasei. J Dairy Sci 89(7):2414–2423
Durmaz EN, Higgins DL, Klaenhammer TR (1992) Molecular characterization of a second abortive phage resistance gene present in Lactococcus lactis subsp. Lactis ME2. J Bacteriol 174(22):7463–7469
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Khoa HV, Midorikawa Y, Uchino T, Nakai T, Kato G, Kondo H, Sano M (2017) Complete genome sequence of the lytic giant bacteriophage pT24 infecting Tenacibaculum spp., isolated from a shrimp culture pond. Genome Announc 5(27):e00081-17
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29(12):2607–2618
Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11(1):119
Marchler-Bauer A et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1):D200–D203
Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J (2008) DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25(1):119–120
Letunic I, Doerks T, Bork P (2014) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43(D1):D257–D260
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964
Möller S, Croning MD, Apweiler R (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17(7):646–653
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
McNair K, Bailey BA, Edwards RA (2012) PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 28(5):614–618
Liu B, Pop M (2009) ARDB—antibiotic resistance genes database. Nucleic Acids Res 37(Suppl 1):D443–D447
Chen L, Xiong Z, Sun L, Yang J, Jin Q (2011) VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res 40(D1):D641–D645
Kokkari C, Sarropoulou E, Bastias R, Mandalakis M, Katharios P (2018) Isolation and characterization of a novel bacteriophage infecting Vibrio alginolyticus. Arch Microbiol 13:1–12
Christensen GD, Simpson WA, Younger JJ, Baddour LM, Burrett FF, Melton DM, Beachey EH (1988) Adherence of coagulase negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of Staphylococci to medical devices. J Clin Microbiol 22(1):996–1006
Stepanovic S, Vukovic D, Hola V, Bonaventura GD, Djukic S, Cirkovic I, Ruzicka F (2007) Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis 115(8):891–899
Meng X, Shi Y, Ji W, Meng X, Zhang J, Wang H, Yan Y (2011) Application of a bacteriophage lysin to disrupt biofilms formed by the animal pathogen, Streptococcus suis. Appl Environ Microbiol 77:8272–8279
Rao BM, Lalitha KV (2015) Bacteriophages for aquaculture: are they beneficial or inimical. Aquaculture 437:146–154
Alagappan KM, Deivasigamani B, Somasundaram ST, Kumaran S (2010) Occurrence of Vibrio parahaemolyticus and its specific phages from shrimp ponds in East Coast of India. Curr Microbiol 61(4):235–240
Stalin N, Srinivasan P (2016) Characterization of Vibrio parahaemolyticus and its specific phage from shrimp pond in Palk Strait, South East Coast of India. Biologicals 44(6):526–533
Singh A, Poshtiban S, Evoy S (2013) Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors (Basel) 13:1763–1786. https://doi.org/10.3390/s130201763
Peltomaa R, Lopez-Perolio I, Benito-Pena E, Barderas R, Moreno-Bondi MC (2016) Application of bacteriophages in sensor development. Anal Bioanal Chem 408:1805–1828. https://doi.org/10.1007/s00216-015-9087-2
Goldberg E, Grinius L, Letellier L (1994) Recognition, attachment, and injection. In: Drake JW, Kreuzer KN, Mosig G, Hall DH, Eiserling FA et al (eds) Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, pp 347–356
Wang N (2006) Lysis timing and bacteriophage fitness. Genetics 172(1):17–26
Kerby GP, Godwy RA, Dillon ES, Dillon ML, Csáky TZ, Sharp DG, Beard JW (1949) Purification, pH stability and sedimentation properties of the T7 bacteriophage of Escherichia coli. J Immunol 63:93–107
Ackermann HW, Tremblay D, Moineau S (2004) Long-term bacteriophage preservation. WFCC Newslett 38:35–40
Feng YY, Ong SL, Hu JY, Tan XL, Ng WJ (2003) Effects of pH and temperature on the survival of coliphages MS2 and Qbeta. J Ind Microbiol Biotechnol 30:549–552
Jończyk E, Kłak M, Międzybrodzki R, Górski A (2011) The influence of external factors on bacteriophages. Folia Microbiol 56(3):191–200
Lal TM, Sano M, Ransangan J (2016) Genome characterization of a novel vibriophage VpKK5 (Siphoviridae) specific to fish pathogenic strain of Vibrio parahaemolyticus. J Basic Microbiol 56(8):872–888
Zhang H, Yang Z, Zhou Y, Bao H, Wang R, Li T, Zhou X (2018) Application of a phage in decontaminating Vibrio parahaemolyticus in oysters. Int J Food Microbiol 275:24–33
Wong HC, Wang TY, Yang CW, Tang CT, Ying C, Wang CH, Chang WH (2018) Characterization of a lytic vibriophage VP06 of Vibrio parahaemolyticus. Res Microbiol
Anany H, Lingohr EJ, Villegas A, Ackermann HW, She YM, Griffiths MW, Kropinski AM (2011) A Shigella boydii bacteriophage which resembles Salmonella phage ViI. Virol J 8(1):242
Kato M, Frick DN, Lee J, Tabor S, Richardson CC, Ellenberger T (2001) A complex of the bacteriophage T7 primase-helicase and DNA polymerase directs primer utilization. J Biol Chem 276(24):21809–21820
Yang H, Ma Y, Wang Y, Yang H, Shen W, Chen X (2014) Transcription regulation mechanisms of bacteriophages: recent advances and future prospects. Bioengineered 5(5):300–304
Kondabagil KR, Rao VB (2006) A critical coiled coil motif in the small terminase, gp16, from bacteriophage T4: insights into DNA packaging initiation and assembly of packaging motor. J Mol Biol 358(1):67–82
Bartual SG, Otero JM, Garcia-Doval C, Llamas-Saiz AL, Kahn R, Fox GC, van Raaij MJ (2010) Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc Natl Acad Sci 107(47):20287–20292
Lee HS, Choi S, Shin H, Lee JH, Choi SH (2014) Vibrio vulnificus bacteriophage SSP002 as a possible biocontrol agent. Appl Environ Microbiol 80(2):515–524
Kao SH, McClain WH (1980) Baseplate protein of bacteriophage T4 with both structural and lytic functions. J Virol 34:95–103
Wang IN, Smith DL, Young R (2000) Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol 54(1):799–825
Oechslin F (2018) Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10(7):351
Brzozowska E, Pyra A, Pawlik K, Górska S, Gamian A (2018) The antibiofilm activity of dual-function tail tubular protein B from KP32 phage
Adams MH, Park BH (1956) An enzyme produced by a phage–host cell system. 11. The properties of the polysaccharide depolymerase. Virology 2:719–736
Acknowledgements
The authors acknowledge the Department of Biotechnology, Cochin University of Science and Technology, for providing all facilities for research. The first author acknowledges DST-PURSE for a Junior Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with animals or human participants performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Matamp, N., Bhat, S.G. Genome characterization of novel lytic Myoviridae bacteriophage ϕVP-1 enhances its applicability against MDR-biofilm-forming Vibrio parahaemolyticus. Arch Virol 165, 387–396 (2020). https://doi.org/10.1007/s00705-019-04493-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04493-6