Abstract
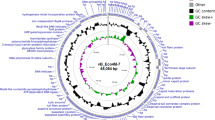
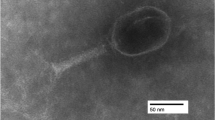
Escherichia coli bacteriophage Gostya9 (genus T5virus) was isolated from horse feces collected in Moscow, Russia, in 2013. This phage was associated in a single plaque with the previously reported phage 9g and was subsequently purified. Analysis of the complete genomic sequence of Gostya9 revealed that it is closely related to the T5-like bacteriophage DT57C, which had been isolated at the same location in 2007. These two viruses share 79.5% nucleotide sequence identity, which is below the 95% threshold applied currently to demarcate bacteriophage species. The most significant features distinguishing Gostya9 from DT57C include 1) the presence of one long tail fiber protein gene, 122c (ltf), instead of the two genes, ltfA and ltfB, that are present in DT57C; 2) the absence of the gene for the receptor-blocking lytic conversion lipoprotein precursor llp; and 3) the divergence of the receptor-recognition protein, pb5, which is only distantly related at the amino acid sequence level. The observed features of the Gostya9 adsorption apparatus are suggestive of a possible novel specificity for the final receptor and make this phage interesting for possible direct application in phage therapy of E. coli infections or as a source of receptor-recognition protein for engineering new phage specificities.


Similar content being viewed by others
References
Adriaenssens E, Brister JR (2017) How to name and classify your phage: an informal guide. Viruses 9:70
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Flayhan A, Wien F, Paternostre M, Boulanger P, Breyton C (2012) New insights into pb5, the receptor binding protein of bacteriophage T5, and its interaction with its Escherichia coli receptor FhuA. Biochimie 94:1982–1989
Golomidova A, Kulikov E, Isaeva A, Manykin A, Letarov A (2007) The diversity of coliphages and coliforms in horse feces reveals a complex pattern of ecological interactions. Appl Environ Microbiol 73:5975–5981
Golomidova AK, Kulikov EE, Prokhorov NS, Guerrero-Ferreira RC, Ksenzenko VN, Tarasyan KK, Letarov AV (2015) Complete genome sequences of T5-related Escherichia coli bacteriophages DT57C and DT571/2 isolated from horse feces. Arch Virol 160:3133–3137
Golomidova AK, Kulikov EE, Prokhorov NS, Guerrero-Ferreira RC, Knirel YA, Kostryukova ES, Tarasyan KK, Letarov AV (2016) Branched lateral tail fiber organization in T5-like bacteriophages DT57C and DT571/2 is revealed by genetic and functional analysis. Viruses 8:26
Heller K, Braun V (1982) Polymannose O-antigens of Escherichia coli, the binding sites for the reversible adsorption of bacteriophage T5+ via the L-shaped tail fibers. J Virol 41:222–227
Heller KJ (1984) Identification of the phage gene for host receptor specificity by analyzing hybrid phages of T5 and BF23. Virology 139:11–21
Killmann H, Videnov G, Jung G, Schwarz H, Braun V (1995) Identification of receptor binding sites by competitive peptide mapping: phages T1, T5, and phi 80 and colicin M bind to the gating loop of FhuA. J Bacteriol 177:694–698
Knirel YA, Prokhorov NS, Shashkov AS, Ovchinnikova OG, Zdorovenko EL, Liu B, Kostryukova ES, Larin AK, Golomidova AK, Letarov AV (2015) Variations in O-antigen biosynthesis and O-acetylation associated with altered phage sensitivity in Escherichia coli 4s. J Bacteriol 197:905–912
Kulikov EE, Golomidova AK, Letarova MA, Kostryukova ES, Zelenin AS, Prokhorov NS, Letarov AV (2014) Genomic sequencing and biological characteristics of a novel Escherichia coli bacteriophage 9g, a putative representative of a new Siphoviridae genus. Viruses 6:5077–5092
Mondigler M, Ayoub AT, Heller KJ (2006) The DNA region of phage BF23 encoding receptor binding protein and receptor blocking lipoprotein lacks homology to the corresponding region of closely related phage T5. J Basic Microbiol 46:116–125
Pedruzzi I, Rosenbusch JP, Locher KP (1998) Inactivation in vitro of the Escherichia coli outer membrane protein FhuA by a phage T5-encoded lipoprotein. FEMS Microbiol Lett 168:119–125
Rabsch W, Ma L, Wiley G, Najar FZ, Kaserer W, Schuerch DW, Klebba JE, Roe BA, Laverde Gomez JA, Schallmey M, Newton SM, Klebba PE (2007) FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. J Bacteriol 189:5658–5674
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Sritha KS, Bhat SG (2018) Genomics of Salmonella phage PhiStp1: candidate bacteriophage for biocontrol. Virus Genes 54:311–318
Sváb D, Falgenhauer L, Rohde M, Szabó J, Chakraborty T, Tóth I (2018) Identification and characterization of T5-like bacteriophages representing two novel subgroups from food products. Front Microbiol 9:202
Wang A, Nizran P, Malone MA, Riley T (2013) Urinary tract infections. Primary Care 40:687–706
Acknowledgements
We are grateful to Prof. Elizabeth M. Kutter from Evergreen State College, Olympia, USA, for critical reading and linguistic corrections of the manuscript. This work was supported by RSF grant no. 15-15-00134P.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All research was conducted in compliance with the national ethical standard code.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Handling Editor: Tim Skern.
Rights and permissions
About this article
Cite this article
Golomidova, A.K., Kulikov, E.E., Babenko, V.V. et al. Escherichia coli bacteriophage Gostya9, representing a new species within the genus T5virus. Arch Virol 164, 879–884 (2019). https://doi.org/10.1007/s00705-018-4113-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-4113-2