Abstract
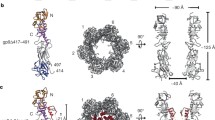
P22 bacteriophage has been studied extensively and has served as a model for many important processes such as in vivo protein folding, protein aggregation and protein-protein interactions. The trimeric tailspike protein (TSP) serves as the receptor-binding protein for the P22 bacteriophage to the bacterial host. The homotrimeric P22 tail consists of three chains of 666aa in which the first 108aa form a trimeric dome-like structure which is called the N-terminal domain (NTD) and is responsible for attachment of the tailspike protein to the rest of the phage particle structure in the phage assembly pathway. Knowledge of this interaction requires information on what amino acids are interacting in the interface and how the NTD structure is maintained. The first 23aa form the “stem peptide” which originates at the dome top and terminates at the dome bottom. It contains a hydrophobic valine patch (V8-V9-V10) located within the dome structure. It is hypothesized that the interaction between the hydrophobic valine patch located on stem peptide and the adjacent polypeptide is critical for the interchain interaction which should be important for the stability of the P22 TSP NTD itself. To test this hypothesis, each amino acid in the valine residues is substituted by an acid, a basic, and a hydrophobic amino acid. The results of such substitutions are presented as well as associated studies. The data strongly suggest that the valine patch is of critical importance in the hydrophobic interaction between stem peptide valine patch and an adjacent chain.





Similar content being viewed by others
References
Nobrega FL, Costa AR, Kluskens LD, Azeredo J (2015) Revisiting phage therapy: new applications for old resources. Trends Microbiol 23:185–191
Prevelige Jr PE (2006) Bacteriophage P22. In: Calendar R (ed) The bacteriophages. Oxford Press, New York, pp. 457–468
Switt AIM, Sulakvelidze A, Weidmann M, Kropinski AM, Wishart DS, Poppe C, Liang Y (2015) Salmonella phages and prophages: genomics, taxonomy, and applied aspects. Methods Mol Biol 1225:237–287
Aksyuk AA, Rossmann MG (2011) Bacteriophage assembly. Viruses 3:172–203
Domitrovic T, Movahed N, Bothner B, Matsui T, Wang Q, Doerschuk P, Johnson J (2013) Virus assembly and maturation: auto-regulation through allosteric molecular switches. J Mol Biol 425:1488–1496
Sauer RT, Krovatin W, Poteete AR, Berget PB (1982) Phage P22 tail protein: gene and amino acid sequence. Biochemistry 21:5811–5815
Seul A, Muller JJ, Andres D, Stettner E, Heinemann U, Seckler R (2014) Bacteriophage P22 tailspike: structure of the complete protein and the function of interdomain linker. Acta Cryst 70:1336–1345
Weigele PR, Scanlon E, King J (2003) Homotrimeric, ß-Stranded viral adhesins and tail proteins. J Bacteriol 185:4022–4030
Goldenberg D, Berget P, King J (1982) Maturation of the tail spike endorhamnosidase of Salmonella Phage P22. J Biol Chem 257:7864–7871
Poteete AR (1988) The Bacteriophage P22. In: Calendar R (ed) The Bacteriophages. Plenum Press, New York, pp 647–682
Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P (1994) Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265:383–386
Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R (1997) Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 Ǻ, fully refined structure of the endorhamnosidase at 1.56 Ǻ resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol 267:865–880
Baxa U, Steinbacher S, Miller S, Weintraub A, Huber R, Seckler R (1996) Interactions of phage P22 tails with their cellular receptors. Salmonella o-Antigen Polysacch Biophys J 71:2040–2048
Palmer C, Williams J, Dean D, Johnson S, Wu H, Jackson D, Villafane R (2014) Stem mutants in the N-terminal domain of the phage P22 tailspike protein. Am J Microbiol Res 2:1–7
Keskin O, Gursoy A, Ma B, Nussinov R (2008) Principles of protein–protein interactions: what are the preferred ways for proteins to interact? Chem Rev 108:1225–1244
Villafane R, Costa S, Ahmed R, Salgado C (2005) Conservation of the N-terminus of some phage tail proteins. Arch Virol 150:2609–2621
Salgado CJ, Zayas M, Villafane R (2004) Homology between two different Salmonella phages: Salmonella enterica serovar Typhimurium phage P22 and Salmonella enterica serovar Anatum var. 15+phage ε34. Virus Genes 29:87–98
Mateu MG (2009) The capsid protein of human immunodeficiency virus: intersubunit interactions during virus assembly. FEBS J 276:6098–6109
Finzi D, Dieffenbach CW, Basavappa R (2007) Defining and solving the essential protein–protein interactions in HIV infections. J Struct Biol 158:148–155
Wang W-H, Chang L-K, Liu S-T (2011) Molecular interactions of Epstein-Barr capsid proteins. J Virol 85:1618–1624
Robinson A, King J (1997) Disulphide-bonded intermediate on the folding and assembly pathway of a non-disulphide bonded protein. Nat Struct Mol Biol 4:450–455
Kamei DT, Liu C, Haase-Pettingell C, King JA, Wang DIC, Blandkschtein D (2002) Understanding viral partitioning in two-phase aqueous nonionic micellar systems: 2. Role of attractive interactions between viruses and micelles. Biotech Bioeng 78:190–202
Schwarz J, Berget P (1989) The isolation and sequence of missense and nonsense mutations in the cloned bacteriophage P22 tailspike protein gene. Genetics 121:635–649
Betts S, Speed M, King J (1999) Detection of early aggregation intermediates by native gel electrophoresis and native western blotting. Methods Enzymol 309:333–350
Roskams J, Rodgers L (2002) Electrophoretic separation of proteins and nucleic acids. In: Roskams J, Rodgers L (eds) Lab Ref: A handbook of recipes, reagents, and other reference tools for use at the bench. Cold Spring Harbor Press, New York, pp 63–91
Maurides PA, Schwarz JJ, Berget PB (1990) Intragenic suppression of a capsid assembly-defective P22 tailspike mutation. Genetics 125:673–681
Chen B, King J (1991) Thermal unfolding pathway for the thermostable P22 tailspike endorhamnosidase. Biochemistry 30:6260–6269
Sturtevant J, Yu M, Haase-Pettingell C, King J (1989) Thermostability of temperature-sensitive folding mutants of the P22 tailspike protein. J Biol Chem 264:10693–10698
Burley S, Petsko G (1985) Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science 229:23–28. https://doi.org/10.1126/science.3892686
Kessel A, Ben-Tal N (2011) Introduction to proteins, 1st edn. CRC Press, Boca Raton, pp 44–58
Keskin O, Gursoy A, Ma B, Nussinov R (2008) Principles of protein–protein interactions: what are the preferred ways for proteins to interact? Chem Rev 108:1225–1244
Prevost M, Wodak S, Tidor B, Karplus M (1991) Contribution of the hydrophobic effect to protein stability: analysis based on simulations of the Ile-96—Ala mutation in barnase. PNAS 88:10880–10884
Manning M, Colón W (2004) Structural basis of protein kinetic stability: resistance to sodium dodecyl sulfate suggests a central role for rigidity and a bias toward β-sheet structure. Biochemistry 43:11248–11254
Xia K, Zhang S, Bathrick B, Liu S, Garcia Y, Colón W (2012) Quantifying the kinetic stability of hyperstable proteins via time-dependent SDS trapping. Biochemistry 51:100–107
Freiberg A, Morona R, Van Den Bosch L, Jung C, Behlke J, Carlin N, Seckler R, Baxa U (2002) The Tailspike Protein of STIgella Phage Sf6: a structural homolog of salmonella phage p22 tailspike protein without sequence similarity in the β-helix domain. J Biol Chem 278(3):1542–1548
Wen J, Arthur K, Chemmalil L, Muzammil S, Gabrielson J, Jiang Y (2012) Applications of can differential scanning calorimetry for thermal stability analysis of proteins: qualification of DSC. J Pharmaceut Sci 101:955–964
Acknowledgements
These studies were supported by the Deanships of the College of Science, Mathematics and Technology at the Alabama State University and by the Department of Chemistry and Biochemistry at Huntingdon College. The authors also thank the anonymous reviewers for insightful comments.
Funding
This work was funded in part by Deanship of the College of Science, Technology, Engineering and Mathematics (C-STEM) from Alabama State University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: T. K. Frey.
Rights and permissions
About this article
Cite this article
Williams, J., Venkatesan, K., Ayariga, J.A. et al. A genetic analysis of an important hydrophobic interaction at the P22 tailspike protein N-terminal domain. Arch Virol 163, 1623–1633 (2018). https://doi.org/10.1007/s00705-018-3777-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3777-y